Fatty Acid Synthase as Interacting Anticancer Target of the Terpenoid Myrianthic Acid Disclosed by MS-Based Proteomics Approaches
- PMID: 38892106
- PMCID: PMC11172900
- DOI: 10.3390/ijms25115918
Fatty Acid Synthase as Interacting Anticancer Target of the Terpenoid Myrianthic Acid Disclosed by MS-Based Proteomics Approaches
Abstract
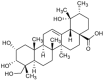
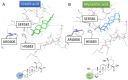
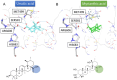
This research focuses on the target deconvolution of the natural compound myrianthic acid, a triterpenoid characterized by an ursane skeleton isolated from the roots of Myrianthus arboreus and from Oenothera maritima Nutt. (Onagraceae), using MS-based chemical proteomic techniques. Application of drug affinity responsive target stability (DARTS) and targeted-limited proteolysis coupled to mass spectrometry (t-LiP-MS) led to the identification of the enzyme fatty acid synthase (FAS) as an interesting macromolecular counterpart of myrianthic acid. This result, confirmed by comparison with the natural ursolic acid, was thoroughly investigated and validated in silico by molecular docking, which gave a precise picture of the interactions in the MA/FAS complex. Moreover, biological assays showcased the inhibitory activity of myrianthic acid against the FAS enzyme, most likely related to its antiproliferative activity towards tumor cells. Given the significance of FAS in specific pathologies, especially cancer, the myrianthic acid structural moieties could serve as a promising reference point to start the potential development of innovative approaches in therapy.
Keywords: DARTS; drug discovery; fatty acid synthase; functional proteomics; molecular docking; preclinical investigations; t-LiP.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

