An Ensemble Classifiers for Improved Prediction of Native-Non-Native Protein-Protein Interaction
- PMID: 38892144
- PMCID: PMC11172808
- DOI: 10.3390/ijms25115957
An Ensemble Classifiers for Improved Prediction of Native-Non-Native Protein-Protein Interaction
Abstract
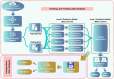
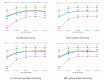
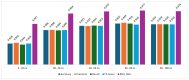
In this study, we present an innovative approach to improve the prediction of protein-protein interactions (PPIs) through the utilization of an ensemble classifier, specifically focusing on distinguishing between native and non-native interactions. Leveraging the strengths of various base models, including random forest, gradient boosting, extreme gradient boosting, and light gradient boosting, our ensemble classifier integrates these diverse predictions using a logistic regression meta-classifier. Our model was evaluated using a comprehensive dataset generated from molecular dynamics simulations. While the gains in AUC and other metrics might seem modest, they contribute to a model that is more robust, consistent, and adaptable. To assess the effectiveness of various approaches, we compared the performance of logistic regression to four baseline models. Our results indicate that logistic regression consistently underperforms across all evaluated metrics. This suggests that it may not be well-suited to capture the complex relationships within this dataset. Tree-based models, on the other hand, appear to be more effective for problems involving molecular dynamics simulations. Extreme gradient boosting (XGBoost) and light gradient boosting (LightGBM) are optimized for performance and speed, handling datasets effectively and incorporating regularizations to avoid over-fitting. Our findings indicate that the ensemble method enhances the predictive capability of PPIs, offering a promising tool for computational biology and drug discovery by accurately identifying potential interaction sites and facilitating the understanding of complex protein functions within biological systems.
Keywords: computational biology; drug discovery; ensemble classifiers; machine learning; protein–protein interaction.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

