Transcriptome Analysis of Potato Leaves under Oxidative Stress
- PMID: 38892181
- PMCID: PMC11172952
- DOI: 10.3390/ijms25115994
Transcriptome Analysis of Potato Leaves under Oxidative Stress
Abstract
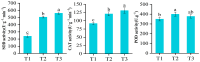
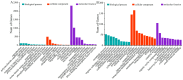
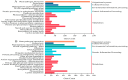
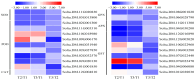
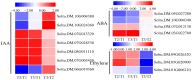
Potato (Solanum tuberosum L.) is a major global food crop, and oxidative stress can significantly impact its growth. Previous studies have shown that its resistance to oxidative stress is mainly related to transcription factors, post-translational modifications, and antioxidant enzymes in vivo, but the specific molecular mechanisms remain unclear. In this study, we analyzed the transcriptome data from potato leaves treated with H2O2 and Methyl viologen (MV), and a control group, for 12 h. We enriched 8334 (CK vs. H2O2) and 4445 (CK vs. MV) differentially expressed genes (DEGs), respectively, and randomly selected 15 DEGs to verify the sequencing data by qRT-PCR. Gene ontology (GO) enrichment analysis showed that the DEGs were mainly concentrated in cellular components and related to molecular function, and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis indicated that most of the DEGs were related to metabolic pathways, plant hormone signal transduction, MAPK-signaling pathway, and plant-pathogen interactions. In addition, several candidate transcription factors, mainly including MYB, WRKY, and genes associated with Ca2+-mediated signal transduction, were also found to be differentially expressed. Among them, the plant hormone genes Soltu.DM.03G022780 and Soltu.DM.06G019360, the CNGC gene Soltu.DM.06G006320, the MYB transcription factors Soltu.DM.06G004450 and Soltu.DM.09G002130, and the WRKY transcription factor Soltu.DM.06G020440 were noticeably highly expressed, which indicates that these are likely to be the key genes in the regulation of oxidative stress tolerance. Overall, these findings lay the foundation for further studies on the molecular mechanisms of potato leaves in response to oxidative stress.
Keywords: Solanum tuberosum L.; differentially expressed genes; oxidative stress; reactive oxygen species; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Martinez C.A., Loureiro M.E., Oliva M.A., Maestri M. Differential responses of superoxide dismutase in freezing resistant Solanum curtilobum and freezing sensitive Solanum tuberosum subjected to oxidative and water stress. Plant Sci. 2001;160:505–515. doi: 10.1016/S0168-9452(00)00418-0. - DOI - PubMed
-
- Jacob S., Dietz K.J. Systematic analysis of superoxide-dependent signaling in plant cells: Usefulness and specificity of methyl viologen application. Astron. Astrophys. Suppl. 2009;136:179–196.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

