A Double-Humanized Mouse Model for Studying Host Gut Microbiome-Immune Interactions in Gulf War Illness
- PMID: 38892281
- PMCID: PMC11172868
- DOI: 10.3390/ijms25116093
A Double-Humanized Mouse Model for Studying Host Gut Microbiome-Immune Interactions in Gulf War Illness
Abstract
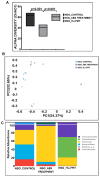
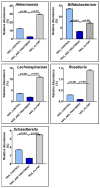
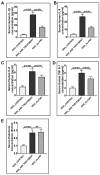
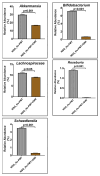
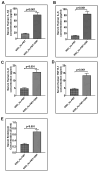
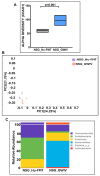
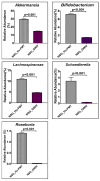
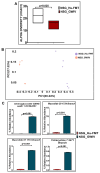
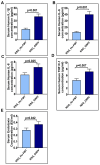
Unraveling the multisymptomatic Gulf War Illness (GWI) pathology and finding an effective cure have eluded researchers for decades. The chronic symptom persistence and limitations for studying the etiologies in mouse models that differ significantly from those in humans pose challenges for drug discovery and finding effective therapeutic regimens. The GWI exposome differs significantly in the study cohorts, and the above makes it difficult to recreate a model closely resembling the GWI symptom pathology. We have used a double engraftment strategy for reconstituting a human immune system coupled with human microbiome transfer to create a humanized-mouse model for GWI. Using whole-genome shotgun sequencing and blood immune cytokine enzyme linked immunosorbent assay (ELISA), we show that our double humanized mice treated with Gulf War (GW) chemicals show significantly altered gut microbiomes, similar to those reported in a Veteran cohort of GWI. The results also showed similar cytokine profiles, such as increased levels of IL-1β, IL-6, and TNF R-1, in the double humanized model, as found previously in a human cohort. Further, a novel GWI Veteran fecal microbiota transfer was used to create a second alternative model that closely resembled the microbiome and immune-system-associated pathology of a GWI Veteran. A GWI Veteran microbiota transplant in humanized mice showed a human microbiome reconstitution and a systemic inflammatory pathology, as reflected by increases in interleukins 1β, 6, 8 (IL-1β, IL-6, IL-8), tumor necrosis factor receptor 1 (TNF R-1), and endotoxemia. In conclusion, though preliminary, we report a novel in vivo model with a human microbiome reconstitution and an engrafted human immune phenotype that may help to better understand gut-immune interactions in GWI.
Keywords: IL-6; NSG; TNF R-1; bacteriome; gut–immune axis; humanized mice.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- White R.F., Steele L., O’Callaghan J.P., Sullivan K., Binns J.H., Golomb B.A., Bloom F.E., Bunker J.A., Crawford F., Graves J.C., et al. Recent research on Gulf War illness and other health problems in veterans of the 1991 Gulf War: Effects of toxicant exposures during deployment. Cortex. 2016;74:449–475. doi: 10.1016/j.cortex.2015.08.022. - DOI - PMC - PubMed
-
- Duong L.M., Nono Djotsa A.B.S., Vahey J., Steele L., Quaden R., Harrington K.M., Ahmed S.T., Polimanti R., Streja E., Gaziano J.M., et al. Association of Gulf War Illness with Characteristics in Deployed vs. Non-Deployed Gulf War Era Veterans in the Cooperative Studies Program 2006/Million Veteran Program 029 Cohort: A Cross-Sectional Analysis. Int. J. Environ. Res. Public Health. 2022;20:258. doi: 10.3390/ijerph20010258. - DOI - PMC - PubMed

