Michael Acceptors as Anti-Cancer Compounds: Coincidence or Causality?
- PMID: 38892287
- PMCID: PMC11172677
- DOI: 10.3390/ijms25116099
Michael Acceptors as Anti-Cancer Compounds: Coincidence or Causality?
Abstract
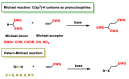
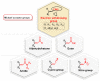
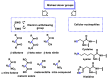
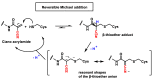
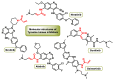
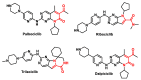
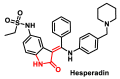
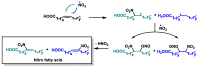
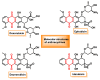
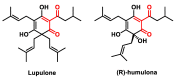
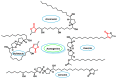
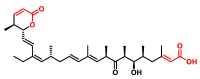
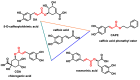
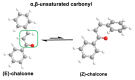
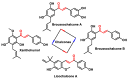
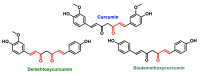
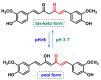
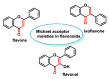
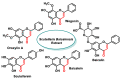
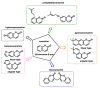
Michael acceptors represent a class of compounds with potential anti-cancer properties. They act by binding to nucleophilic sites in biological molecules, thereby disrupting cancer cell function and inducing cell death. This mode of action, as well as their ability to be modified and targeted, makes them a promising avenue for advancing cancer therapy. We are investigating the molecular mechanisms underlying Michael acceptors and their interactions with cancer cells, in particular their ability to interfere with cellular processes and induce apoptosis. The anti-cancer properties of Michael acceptors are not accidental but are due to their chemical structure and reactivity. The electrophilic nature of these compounds allows them to selectively target nucleophilic residues on disease-associated proteins, resulting in significant therapeutic benefits and minimal toxicity in various diseases. This opens up new perspectives for the development of more effective and precise cancer drugs. Nevertheless, further studies are essential to fully understand the impact of our discoveries and translate them into clinical practice.
Keywords: Michael acceptor compounds; chemotherapy; neoplastic metabolism.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
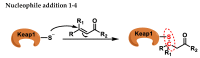


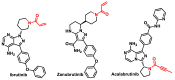
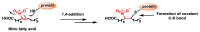
Similar articles
-
The Emerging Therapeutic Potential of Nitro Fatty Acids and Other Michael Acceptor-Containing Drugs for the Treatment of Inflammation and Cancer.Front Pharmacol. 2020 Sep 3;11:1297. doi: 10.3389/fphar.2020.01297. eCollection 2020. Front Pharmacol. 2020. PMID: 33013366 Free PMC article. Review.
-
Role of Bcl-2 in tumour cell survival and implications for pharmacotherapy.J Pharm Pharmacol. 2012 Dec;64(12):1695-702. doi: 10.1111/j.2042-7158.2012.01526.x. Epub 2012 Apr 25. J Pharm Pharmacol. 2012. PMID: 23146031 Review.
-
Identification of Michael acceptor-centric pharmacophores with substituents that yield strong thioredoxin reductase inhibitory character correlated to antiproliferative activity.Antioxid Redox Signal. 2013 Oct 10;19(11):1149-65. doi: 10.1089/ars.2012.4909. Epub 2013 Feb 28. Antioxid Redox Signal. 2013. PMID: 23311917 Free PMC article.
-
New drugs in cancer therapy, National Tumor Institute, Naples, 17-18 June 2004.Anticancer Drugs. 2005 Feb;16(2):211-21. doi: 10.1097/00001813-200502000-00014. Anticancer Drugs. 2005. PMID: 15655420
-
Role of Thiol Reactivity for Targeting Mutant p53.Cell Chem Biol. 2018 Oct 18;25(10):1219-1230.e3. doi: 10.1016/j.chembiol.2018.06.013. Epub 2018 Jul 26. Cell Chem Biol. 2018. PMID: 30057300
Cited by
-
Dual-Action Therapeutics: DNA Alkylation and Antimicrobial Peptides for Cancer Therapy.Cancers (Basel). 2024 Sep 10;16(18):3123. doi: 10.3390/cancers16183123. Cancers (Basel). 2024. PMID: 39335095 Free PMC article. Review.
-
Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Human 20S Proteasome Inhibitors: Extension to the Prime Site.ACS Omega. 2025 Jul 16;10(29):31549-31567. doi: 10.1021/acsomega.5c02128. eCollection 2025 Jul 29. ACS Omega. 2025. PMID: 40757351 Free PMC article.
-
Antimicrobial and ADME properties of methoxylated, methylated and nitrated 2-hydroxynaphthalene-1 carboxanilides.ADMET DMPK. 2025 Feb 8;13(1):2642. doi: 10.5599/admet.2642. eCollection 2025. ADMET DMPK. 2025. PMID: 40161889 Free PMC article.
-
In Silico and In Vitro Analyses of Strawberry-Derived Extracts in Relation to Key Compounds' Metabolic and Anti-Tumor Effects.Int J Mol Sci. 2025 Apr 8;26(8):3492. doi: 10.3390/ijms26083492. Int J Mol Sci. 2025. PMID: 40331930 Free PMC article.
-
Cysteine Alkylation in Enzymes and Transcription Factors: A Therapeutic Strategy for Cancer.Cancers (Basel). 2025 Jun 3;17(11):1876. doi: 10.3390/cancers17111876. Cancers (Basel). 2025. PMID: 40507356 Free PMC article. Review.
References
-
- Su M.-G., Weng J.T.-Y., Hsu J.B.-K., Huang K.-Y., Chi Y.-H., Lee T.-Y. Investigation and identification of functional post-translational modification sites associated with drug binding and protein-protein interactions. BMC Syst. Biol. 2017;11:132. doi: 10.1186/s12918-017-0506-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

