Genome-Wide Identification, Characterization, Evolutionary Analysis, and Expression Pattern of the GPAT Gene Family in Barley and Functional Analysis of HvGPAT18 under Abiotic Stress
- PMID: 38892304
- PMCID: PMC11172788
- DOI: 10.3390/ijms25116101
Genome-Wide Identification, Characterization, Evolutionary Analysis, and Expression Pattern of the GPAT Gene Family in Barley and Functional Analysis of HvGPAT18 under Abiotic Stress
Abstract
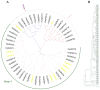
Glycerol-3-phosphoacyltransferase (GPAT) is an important rate-limiting enzyme in the biosynthesis of triacylglycerol (TAG), which is of great significance for plant growth, development, and response to abiotic stress. Although the characteristics of GPAT have been studied in many model plants, little is known about its expression profile and function in barley, especially under abiotic stress. In this study, 22 GPAT genes were identified in the barley genome and divided into three groups (I, II, III), with the latter Group III subdivided further into three subgroups based on the phylogenetic analysis. The analyses of conserved motifs, gene structures, and the three-dimensional structure of HvGPAT proteins also support this classification. Through evolutionary analysis, we determined that HvGPATs in Group I were the earliest to diverge during 268.65 MYA, and the differentiation of other HvGPATs emerged during 86.83-169.84 MYA. The tissue expression profile showed that 22 HvGPAT genes were almost not expressed in INF1 (inflorescence 1). Many functional elements related to stress responses and hormones in cis-element analysis, as well as qRT-PCR results, confirm that these HvGPAT genes were involved in abiotic stress responses. The expression level of HvGPAT18 was significantly increased under abiotic stress and its subcellular localization indicated its function in the endoplasmic reticulum. Various physiological traits under abiotic stress were evaluated using transgenic Arabidopsis to gain further insight into the role of HvGPAT18, and it was found that transgenic seedlings have stronger resistance under abiotic stress than to the wild-type (WT) plants. Overall, our results provide new insights into the evolution and function of the barley GPAT gene family and enable us to explore the molecular mechanism of functional diversity behind the evolutionary history of these genes.
Keywords: Hordeum vulgare L.; abiotic stress; classification; evolution; expression pattern; functional analysis; gene family; glycerol-3-phosphate acyltransferase (GPAT).
Conflict of interest statement
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

