Whole-Genome Sequencing and Analysis of Tumour-Forming Radish (Raphanus sativus L.) Line
- PMID: 38892425
- PMCID: PMC11172632
- DOI: 10.3390/ijms25116236
Whole-Genome Sequencing and Analysis of Tumour-Forming Radish (Raphanus sativus L.) Line
Abstract
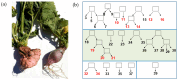
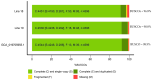
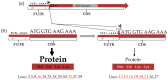
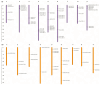
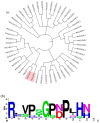
Spontaneous tumour formation in higher plants can occur in the absence of pathogen invasion, depending on the plant genotype. Spontaneous tumour formation on the taproots is consistently observed in certain inbred lines of radish (Raphanus sativus var. radicula Pers.). In this paper, using Oxford Nanopore and Illumina technologies, we have sequenced the genomes of two closely related radish inbred lines that differ in their ability to spontaneously form tumours. We identified a large number of single nucleotide variants (amino acid substitutions, insertions or deletions, SNVs) that are likely to be associated with the spontaneous tumour formation. Among the genes involved in the trait, we have identified those that regulate the cell cycle, meristem activity, gene expression, and metabolism and signalling of phytohormones. After identifying the SNVs, we performed Sanger sequencing of amplicons corresponding to SNV-containing regions to validate our results. We then checked for the presence of SNVs in other tumour lines of the radish genetic collection and found the ERF118 gene, which had the SNVs in the majority of tumour lines. Furthermore, we performed the identification of the CLAVATA3/ESR (CLE) and WUSCHEL (WOX) genes and, as a result, identified two unique radish CLE genes which probably encode proteins with multiple CLE domains. The results obtained provide a basis for investigating the mechanisms of plant tumour formation and also for future genetic and genomic studies of radish.
Keywords: CLE; Raphanus sativus; WOX; genomic sequence; inbred lines; single nucleotide variants; spontaneous tumours.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Identification, expression, and functional analysis of CLE genes in radish (Raphanus sativus L.) storage root.BMC Plant Biol. 2016 Jan 27;16 Suppl 1(Suppl 1):7. doi: 10.1186/s12870-015-0687-y. BMC Plant Biol. 2016. PMID: 26821718 Free PMC article.
-
De novo assembly and characterization of the complete chloroplast genome of radish (Raphanus sativus L.).Gene. 2014 Nov 1;551(1):39-48. doi: 10.1016/j.gene.2014.08.038. Epub 2014 Aug 21. Gene. 2014. PMID: 25151309
-
Initiation of spontaneous tumors in radish (Raphanus sativus): Cellular, molecular and physiological events.J Plant Physiol. 2015 Jan 15;173:97-104. doi: 10.1016/j.jplph.2014.07.030. Epub 2014 Sep 18. J Plant Physiol. 2015. PMID: 25462083
-
Draft sequences of the radish (Raphanus sativus L.) genome.DNA Res. 2014 Oct;21(5):481-90. doi: 10.1093/dnares/dsu014. Epub 2014 May 16. DNA Res. 2014. PMID: 24848699 Free PMC article.
-
Genome-wide identification and characterization of NBS-encoding genes in Raphanus sativus L. and their roles related to Fusarium oxysporum resistance.BMC Plant Biol. 2021 Jan 18;21(1):47. doi: 10.1186/s12870-020-02803-8. BMC Plant Biol. 2021. PMID: 33461498 Free PMC article.
Cited by
-
Cellular and Molecular Regulatory Signals in Root Growth and Development.Int J Mol Sci. 2025 Apr 6;26(7):3426. doi: 10.3390/ijms26073426. Int J Mol Sci. 2025. PMID: 40244272 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

