Genome-Wide Identification, Characterisation, and Evolution of the Transcription Factor WRKY in Grapevine (Vitis vinifera): New View and Update
- PMID: 38892428
- PMCID: PMC11172563
- DOI: 10.3390/ijms25116241
Genome-Wide Identification, Characterisation, and Evolution of the Transcription Factor WRKY in Grapevine (Vitis vinifera): New View and Update
Abstract
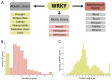
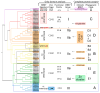
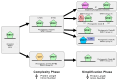
WRKYs are a multigenic family of transcription factors that are plant-specific and involved in the regulation of plant development and various stress response processes. However, the evolution of WRKY genes is not fully understood. This family has also been incompletely studied in grapevine, and WRKY genes have been named with different numbers in different studies, leading to great confusion. In this work, 62 Vitis vinifera WRKY genes were identified based on six genomes of different cultivars. All WRKY genes were numbered according to their chromosomal location, and a complete revision of the numbering was performed. Amino acid variability between different cultivars was assessed for the first time and was greater than 5% for some WRKYs. According to the gene structure, all WRKYs could be divided into two groups: more exons/long length and fewer exons/short length. For the first time, some chimeric WRKY genes were found in grapevine, which may play a specific role in the regulation of different processes: VvWRKY17 (an N-terminal signal peptide region followed by a non-cytoplasmic domain) and VvWRKY61 (Frigida-like domain). Five phylogenetic clades A-E were revealed and correlated with the WRKY groups (I, II, III). The evolution of WRKY was studied, and we proposed a WRKY evolution model where there were two dynamic phases of complexity and simplification in the evolution of WRKY.
Keywords: Vitis vinifera; WRKY transcription factor; genome-wide analyses; grape; grape cultivars; phylogeny.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Cai R., Zhao Y., Wang Y., Lin Y., Peng X., Li Q., Chang Y., Jiang H., Xiang Y., Cheng B. Overexpression of a Maize WRKY58 Gene Enhances Drought and Salt Tolerance in Transgenic Rice. Plant Cell Tissue Organ Cult. (PCTOC) 2014;119:565–577. doi: 10.1007/s11240-014-0556-7. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

