Exploring the Ocular Surface Microbiome and Tear Proteome in Glaucoma
- PMID: 38892444
- PMCID: PMC11172891
- DOI: 10.3390/ijms25116257
Exploring the Ocular Surface Microbiome and Tear Proteome in Glaucoma
Abstract
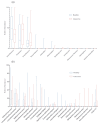
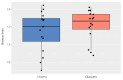
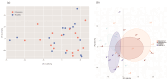
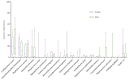
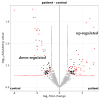
Although glaucoma is a leading cause of irreversible blindness worldwide, its pathogenesis is incompletely understood, and intraocular pressure (IOP) is the only modifiable risk factor to target the disease. Several associations between the gut microbiome and glaucoma, including the IOP, have been suggested. There is growing evidence that interactions between microbes on the ocular surface, termed the ocular surface microbiome (OSM), and tear proteins, collectively called the tear proteome, may also play a role in ocular diseases such as glaucoma. This study aimed to find characteristic features of the OSM and tear proteins in patients with glaucoma. The whole-metagenome shotgun sequencing of 32 conjunctival swabs identified Actinobacteria, Firmicutes, and Proteobacteria as the dominant phyla in the cohort. The species Corynebacterium mastitidis was only found in healthy controls, and their conjunctival microbiomes may be enriched in genes of the phospholipase pathway compared to glaucoma patients. Despite these minor differences in the OSM, patients showed an enrichment of many tear proteins associated with the immune system compared to controls. In contrast to the OSM, this emphasizes the role of the proteome, with a potential involvement of immunological processes in glaucoma. These findings may contribute to the design of new therapeutic approaches targeting glaucoma and other associated diseases.
Keywords: glaucoma; liquid chromatography–tandem mass spectrometry; ocular surface microbiome; tear proteome; whole-metagenome shotgun sequencing.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. Throughout the entirety of the work, ChatGPT 3.5 was utilized for structural assistance in writing the paper, as well as for linguistic and formal revisions.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

