EGFRvIII Confers Sensitivity to Saracatinib in a STAT5-Dependent Manner in Glioblastoma
- PMID: 38892466
- PMCID: PMC11172708
- DOI: 10.3390/ijms25116279
EGFRvIII Confers Sensitivity to Saracatinib in a STAT5-Dependent Manner in Glioblastoma
Abstract
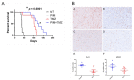
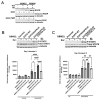
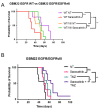
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults, with few effective treatments. EGFR alterations, including expression of the truncated variant EGFRvIII, are among the most frequent genomic changes in these tumors. EGFRvIII is known to preferentially signal through STAT5 for oncogenic activation in GBM, yet targeting EGFRvIII has yielded limited clinical success to date. In this study, we employed patient-derived xenograft (PDX) models expressing EGFRvIII to determine the key points of therapeutic vulnerability within the EGFRvIII-STAT5 signaling axis in GBM. Our findings reveal that exogenous expression of paralogs STAT5A and STAT5B augments cell proliferation and that inhibition of STAT5 phosphorylation in vivo improves overall survival in combination with temozolomide (TMZ). STAT5 phosphorylation is independent of JAK1 and JAK2 signaling, instead requiring Src family kinase (SFK) activity. Saracatinib, an SFK inhibitor, attenuates phosphorylation of STAT5 and preferentially sensitizes EGFRvIII+ GBM cells to undergo apoptotic cell death relative to wild-type EGFR. Constitutively active STAT5A or STAT5B mitigates saracatinib sensitivity in EGFRvIII+ cells. In vivo, saracatinib treatment decreased survival in mice bearing EGFR WT tumors compared to the control, yet in EGFRvIII+ tumors, treatment with saracatinib in combination with TMZ preferentially improves survival.
Keywords: cell signaling; glioblastoma; precision oncology; receptor tyrosine kinase.
Conflict of interest statement
Author Landon Inge was employed by the company Ventana Medical Systems, Roche Diagnostics. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Sarkaria J.N., Hu L.S., Parney I.F., Pafundi D.H., Brinkmann D.H., Laack N.N., Giannini C., Burns T.C., Kizilbash S.H., Laramy J.K., et al. Is the blood–brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro-Oncology. 2017;20:184–191. doi: 10.1093/neuonc/nox175. - DOI - PMC - PubMed
-
- Blomquist M.R., Ensign S.F., D’angelo F., Phillips J.J., Ceccarelli M., Peng S., Halperin R.F., Caruso F.P., Garofano L., Byron S.A., et al. Temporospatial genomic profiling in glioblastoma identifies commonly altered core pathways underlying tumor progression. Neuro-Oncol. Adv. 2020;2:vdaa078. doi: 10.1093/noajnl/vdaa078. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

