Arrestins: A Small Family of Multi-Functional Proteins
- PMID: 38892473
- PMCID: PMC11173308
- DOI: 10.3390/ijms25116284
Arrestins: A Small Family of Multi-Functional Proteins
Abstract
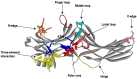
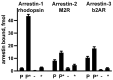
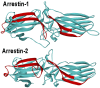
The first member of the arrestin family, visual arrestin-1, was discovered in the late 1970s. Later, the other three mammalian subtypes were identified and cloned. The first described function was regulation of G protein-coupled receptor (GPCR) signaling: arrestins bind active phosphorylated GPCRs, blocking their coupling to G proteins. It was later discovered that receptor-bound and free arrestins interact with numerous proteins, regulating GPCR trafficking and various signaling pathways, including those that determine cell fate. Arrestins have no enzymatic activity; they function by organizing multi-protein complexes and localizing their interaction partners to particular cellular compartments. Today we understand the molecular mechanism of arrestin interactions with GPCRs better than the mechanisms underlying other functions. However, even limited knowledge enabled the construction of signaling-biased arrestin mutants and extraction of biologically active monofunctional peptides from these multifunctional proteins. Manipulation of cellular signaling with arrestin-based tools has research and likely therapeutic potential: re-engineered proteins and their parts can produce effects that conventional small-molecule drugs cannot.
Keywords: GPCR; MAP kinase; arrestin; microtubules; ubiquitin ligase.
Conflict of interest statement
The author declares no conflicts of interest.
Figures
Similar articles
-
Plethora of functions packed into 45 kDa arrestins: biological implications and possible therapeutic strategies.Cell Mol Life Sci. 2019 Nov;76(22):4413-4421. doi: 10.1007/s00018-019-03272-5. Epub 2019 Aug 17. Cell Mol Life Sci. 2019. PMID: 31422444 Free PMC article. Review.
-
Arrestins: Introducing Signaling Bias Into Multifunctional Proteins.Prog Mol Biol Transl Sci. 2018;160:47-61. doi: 10.1016/bs.pmbts.2018.07.007. Epub 2018 Sep 6. Prog Mol Biol Transl Sci. 2018. PMID: 30470292 Free PMC article. Review.
-
β-arrestins and G protein-coupled receptor trafficking.Handb Exp Pharmacol. 2014;219:173-86. doi: 10.1007/978-3-642-41199-1_9. Handb Exp Pharmacol. 2014. PMID: 24292830 Free PMC article. Review.
-
Receptor sequestration in response to β-arrestin-2 phosphorylation by ERK1/2 governs steady-state levels of GPCR cell-surface expression.Proc Natl Acad Sci U S A. 2015 Sep 15;112(37):E5160-8. doi: 10.1073/pnas.1508836112. Epub 2015 Aug 31. Proc Natl Acad Sci U S A. 2015. PMID: 26324936 Free PMC article.
-
Overview of different mechanisms of arrestin-mediated signaling.Curr Protoc Pharmacol. 2014 Dec 1;67:2.10.1-2.10.9. doi: 10.1002/0471141755.ph0210s67. Curr Protoc Pharmacol. 2014. PMID: 25446289 Free PMC article. Review.
Cited by
-
A small molecule enhances arrestin-3 binding to the β2-adrenergic receptor.bioRxiv [Preprint]. 2024 Dec 13:2024.12.12.628161. doi: 10.1101/2024.12.12.628161. bioRxiv. 2024. Update in: Commun Chem. 2025 Jul 1;8(1):194. doi: 10.1038/s42004-025-01581-4. PMID: 39713392 Free PMC article. Updated. Preprint.
-
A small molecule enhances arrestin-3 binding to the β2-adrenergic receptor.Commun Chem. 2025 Jul 1;8(1):194. doi: 10.1038/s42004-025-01581-4. Commun Chem. 2025. PMID: 40593109 Free PMC article.
-
The Role of Individual Residues in the N-Terminus of Arrestin-1 in Rhodopsin Binding.Int J Mol Sci. 2025 Jan 16;26(2):715. doi: 10.3390/ijms26020715. Int J Mol Sci. 2025. PMID: 39859432 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

