Distinct Gut Microbiota and Arachidonic Acid Metabolism in Obesity-Prone and Obesity-Resistant Mice with a High-Fat Diet
- PMID: 38892512
- PMCID: PMC11174461
- DOI: 10.3390/nu16111579
Distinct Gut Microbiota and Arachidonic Acid Metabolism in Obesity-Prone and Obesity-Resistant Mice with a High-Fat Diet
Abstract
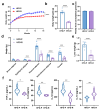
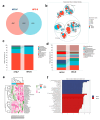
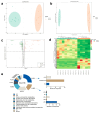
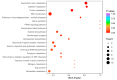
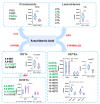
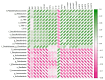
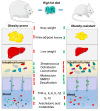
An imbalance of energy intake and expenditure is commonly considered as the fundamental cause of obesity. However, individual variations in susceptibility to obesity do indeed exist in both humans and animals, even among those with the same living environments and dietary intakes. To further explore the potential influencing factors of these individual variations, male C57BL/6J mice were used for the development of obesity-prone and obesity-resistant mice models and were fed high-fat diets for 16 weeks. Compared to the obesity-prone mice, the obesity-resistant group showed a lower body weight, liver weight, adipose accumulation and pro-inflammatory cytokine levels. 16S rRNA sequencing, which was conducted for fecal microbiota analysis, found that the fecal microbiome's structural composition and biodiversity had changed in the two groups. The genera Allobaculumbiota, SMB53, Desulfovibrio and Clostridium increased in the obesity-prone mice, and the genera Streptococcus, Odoribacter and Leuconostoc were enriched in the obesity-resistant mice. Using widely targeted metabolomics analysis, 166 differential metabolites were found, especially those products involved in arachidonic acid (AA) metabolism, which were significantly reduced in the obesity-resistant mice. Moreover, KEGG pathway analysis exhibited that AA metabolism was the most enriched pathway. Significantly altered bacteria and obesity-related parameters, as well as AA metabolites, exhibited strong correlations. Overall, the phenotypes of the obesity-prone and obesity-resistant mice were linked to gut microbiota and AA metabolism, providing new insight for developing an in-depth understanding of the driving force of obesity resistance and a scientific reference for the targeted prevention and treatment of obesity.
Keywords: arachidonic acid metabolism; gut microbiota; obesity prone; obesity resistant; widely targeted metabolomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Overstreet A.C., Grayson B.E., Boger A., Bakke D., Carmody E.M., Bales C.E., Paski S.C., Murphy S.F., Dethlefs C.R., Shannon K.J., et al. Gastrokine-1, an anti-amyloidogenic protein secreted by the stomach, regulates diet-induced obesity. Sci. Rep. 2021;11:9477. doi: 10.1038/s41598-021-88928-8. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

