Overlapping Stromal Alterations in Myeloid and Lymphoid Neoplasms
- PMID: 38893194
- PMCID: PMC11171322
- DOI: 10.3390/cancers16112071
Overlapping Stromal Alterations in Myeloid and Lymphoid Neoplasms
Abstract
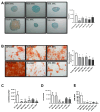
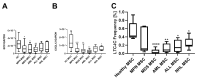
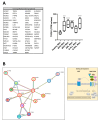
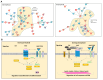
Myeloid and lymphoid neoplasms share the characteristics of potential bone marrow infiltration as a primary or secondary effect, which readily leads to hematopoietic insufficiency. The mechanisms by which clonal malignant cells inhibit normal hematopoietic stem and progenitor cells (HSPCs) in the bone marrow (BM) have not been unraveled so far. Given the pivotal role of mesenchymal stromal cells (MSCs) in the regulation of hematopoiesis in the BM niche it is assumed that MSCs also play a relevant role in the pathogenesis of hematological neoplasms. We aimed to identify overlapping mechanisms in MSCs derived from myeloid and lymphoid neoplasms contributing to disease progression and suppression of HSPCs to develop interventions that target these mechanisms. MSCs derived from healthy donors (n = 44) and patients diagnosed with myeloproliferative neoplasia (n = 11), myelodysplastic syndromes (n = 16), or acute myeloid leukemia (n = 25) and B-Non-Hodgkin lymphoma (n = 9) with BM infiltration and acute lymphoblastic leukemia (n = 9) were analyzed for their functionality and by RNA sequencing. A reduced growth and differentiation capacity of MSCs was found in all entities. RNA sequencing distinguished both groups but clearly showed overlapping differentially expressed genes, including major players in the BMP/TGF and WNT-signaling pathway which are crucial for growth, osteogenesis, and hematopoiesis. Functional alterations in healthy MSCs were inducible by exposure to supernatants from malignant cells, implicating the involvement of these factors in disease progression. Overall, we were able to identify overlapping factors that pose potential future therapeutic targets.
Keywords: ALL; AML; MDS; MPN; MSC; NHL; RNA sequencing; bone marrow microenvironment; hematopoietic insufficiency; lymphoid neoplasms; myeloid neoplasms; osteogenesis.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest The author Felix Bormann is affiliated with Bioinformatics.Expert UG but has no potential interest relationship. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Figures







References
-
- Alaggio R., Amador C., Anagnostopoulos I., Attygalle A.D., Araujo I.B.O., Berti E., Bhagat G., Borges A.M., Boyer D., Calaminici M., et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia. 2022;36:1720–1748. doi: 10.1038/s41375-022-01620-2. - DOI - PMC - PubMed
-
- Arber D.A., Orazi A., Hasserjian R.P., Borowitz M.J., Calvo K.R., Kvasnicka H.M., Wang S.A., Bagg A., Barbui T., Branford S., et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood. 2022;140:1200–1228. doi: 10.1182/blood.2022015850. - DOI - PMC - PubMed
-
- Will B., Zhou L., Vogler T.O., Ben-Neriah S., Schinke C., Tamari R., Yu Y., Bhagat T.D., Bhattacharyya S., Barreyro L., et al. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 2012;120:2076–2086. doi: 10.1182/blood-2011-12-399683. - DOI - PMC - PubMed
-
- Borkhardt A., Wuchter C., Viehmann S., Pils S., Teigler-Schlegel A., Stanulla M., Zimmermann M., Ludwig W.D., Janka-Schaub G., Schrappe M., et al. Infant acute lymphoblastic leukemia—Combined cytogenetic, immunophenotypical and molecular analy-sis of 77 cases. Leukemia. 2002;16:1685–1690. doi: 10.1038/sj.leu.2402595. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

