Exploring the Therapeutic Potential of Petiveria alliacea L. Phytochemicals: A Computational Study on Inhibiting SARS-CoV-2's Main Protease (Mpro)
- PMID: 38893400
- PMCID: PMC11173994
- DOI: 10.3390/molecules29112524
Exploring the Therapeutic Potential of Petiveria alliacea L. Phytochemicals: A Computational Study on Inhibiting SARS-CoV-2's Main Protease (Mpro)
Abstract
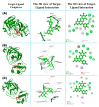
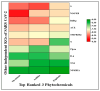
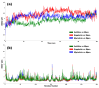
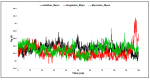
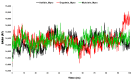
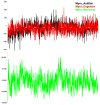
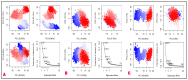
The outbreak of SARS-CoV-2, also known as the COVID-19 pandemic, is still a critical risk factor for both human life and the global economy. Although, several promising therapies have been introduced in the literature to inhibit SARS-CoV-2, most of them are synthetic drugs that may have some adverse effects on the human body. Therefore, the main objective of this study was to carry out an in-silico investigation into the medicinal properties of Petiveria alliacea L. (P. alliacea L.)-mediated phytocompounds for the treatment of SARS-CoV-2 infections since phytochemicals have fewer adverse effects compared to synthetic drugs. To explore potential phytocompounds from P. alliacea L. as candidate drug molecules, we selected the infection-causing main protease (Mpro) of SARS-CoV-2 as the receptor protein. The molecular docking analysis of these receptor proteins with the different phytocompounds of P. alliacea L. was performed using AutoDock Vina. Then, we selected the three top-ranked phytocompounds (myricitrin, engeletin, and astilbin) as the candidate drug molecules based on their highest binding affinity scores of -8.9, -8.7 and -8.3 (Kcal/mol), respectively. Then, a 100 ns molecular dynamics (MD) simulation study was performed for their complexes with Mpro using YASARA software, computed RMSD, RMSF, PCA, DCCM, MM/PBSA, and free energy landscape (FEL), and found their almost stable binding performance. In addition, biological activity, ADME/T, DFT, and drug-likeness analyses exhibited the suitable pharmacokinetics properties of the selected phytocompounds. Therefore, the results of this study might be a useful resource for formulating a safe treatment plan for SARS-CoV-2 infections after experimental validation in wet-lab and clinical trials.
Keywords: SARS-CoV-2 infections; computational approaches; main protease; phytocompound; toxicity.
Conflict of interest statement
All the authors declare no conflicts of interest.
Figures



References
-
- Verma D., Mitra D., Paul M., Chaudhary P., Kamboj A., Thatoi H., Janmeda P., Jain D., Panneerselvam P., Shrivastav R., et al. Potential inhibitors of SARS-CoV-2 (COVID-19) proteases PLpro and Mpro/3CLpro: Molecular docking and simulation studies of three pertinent medicinal plant natural components. Curr. Res. Pharmacol. Drug Discov. 2021;2:100038. doi: 10.1016/j.crphar.2021.100038. - DOI - PMC - PubMed
-
- Sharma P., Joshi T., Mathpal S., Joshi T., Pundir H., Chandra S., Tamta S. Identification of natural inhibitors against Mpro of SARS-CoV-2 by molecular docking, molecular dynamics simulation, and MM/PBSA methods. J. Biomol. Struct. Dyn. 2022;40:2757–2768. doi: 10.1080/07391102.2020.1842806. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

