Structural Analysis of the Large Stokes Shift Red Fluorescent Protein tKeima
- PMID: 38893454
- PMCID: PMC11173989
- DOI: 10.3390/molecules29112579
Structural Analysis of the Large Stokes Shift Red Fluorescent Protein tKeima
Abstract
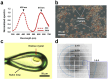
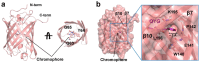
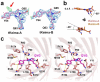
The Keima family comprises large Stokes shift fluorescent proteins that are useful for dual-color fluorescence cross-correlation spectroscopy and multicolor imaging. The tKeima is a tetrameric large Stokes shift fluorescent protein and serves as the ancestor fluorescent protein for both dKeima and mKeima. The spectroscopic properties of tKeima have been previously reported; however, its structural basis and molecular properties have not yet been elucidated. In this study, we present the crystallographic results of the large Stokes shift fluorescent protein tKeima. The purified tKeima protein spontaneously crystallized after purification without further crystallization. The crystal structure of tKeima was determined at 3.0 Å resolution, revealing a β-barrel fold containing the Gln-Tyr-Gly chromophores mainly with cis-conformation. The tetrameric interfaces of tKeima were stabilized by numerous hydrogen bonds and salt-bridge interactions. These key residues distinguish the substituted residues in dKeima and mKeima. The key structure-based residues involved in the tetramer formation of tKeima provide insights into the generation of a new type of monomeric mKeima. This structural analysis expands our knowledge of the Keima family and provides insights into its protein engineering.
Keywords: Keima; fluorescent protein; large Stoke shift; structure; tKeima.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Benaissa H., Ounoughi K., Aujard I., Fischer E., Goïame R., Nguyen J., Tebo A.G., Li C., Le Saux T., Bertolin G., et al. Engineering of a fluorescent chemogenetic reporter with tunable color for advanced live-cell imaging. Nat. Commun. 2021;12:6989. doi: 10.1038/s41467-021-27334-0. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

