Decoding the κ Opioid Receptor (KOR): Advancements in Structural Understanding and Implications for Opioid Analgesic Development
- PMID: 38893511
- PMCID: PMC11173883
- DOI: 10.3390/molecules29112635
Decoding the κ Opioid Receptor (KOR): Advancements in Structural Understanding and Implications for Opioid Analgesic Development
Abstract
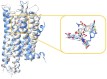
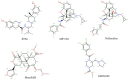
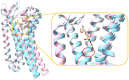
The opioid crisis in the United States is a significant public health issue, with a nearly threefold increase in opioid-related fatalities between 1999 and 2014. In response to this crisis, society has made numerous efforts to mitigate its impact. Recent advancements in understanding the structural intricacies of the κ opioid receptor (KOR) have improved our knowledge of how opioids interact with their receptors, triggering downstream signaling pathways that lead to pain relief. This review concentrates on the KOR, offering crucial structural insights into the binding mechanisms of both agonists and antagonists to the receptor. Through comparative analysis of the atomic details of the binding site, distinct interactions specific to agonists and antagonists have been identified. These insights not only enhance our understanding of ligand binding mechanisms but also shed light on potential pathways for developing new opioid analgesics with an improved risk-benefit profile.
Keywords: agonist; antagonist; binding; ligand; mechanism; opioid; receptor; structure.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Molecular Interaction Between Butorphanol and κ-Opioid Receptor.Anesth Analg. 2020 Sep;131(3):935-942. doi: 10.1213/ANE.0000000000005017. Anesth Analg. 2020. PMID: 32701545 Free PMC article.
-
Chemotype-selective modes of action of κ-opioid receptor agonists.J Biol Chem. 2013 Nov 29;288(48):34470-83. doi: 10.1074/jbc.M113.515668. Epub 2013 Oct 11. J Biol Chem. 2013. PMID: 24121503 Free PMC article.
-
Development of Diphenethylamines as Selective Kappa Opioid Receptor Ligands and Their Pharmacological Activities.Molecules. 2020 Nov 2;25(21):5092. doi: 10.3390/molecules25215092. Molecules. 2020. PMID: 33147885 Free PMC article. Review.
-
Characterization of kappa opioid receptor mediated, dynorphin-stimulated [35S]GTPγS binding in mouse striatum for the evaluation of selective KOR ligands in an endogenous setting.Neuropharmacology. 2015 Dec;99:131-41. doi: 10.1016/j.neuropharm.2015.07.001. Epub 2015 Jul 6. Neuropharmacology. 2015. PMID: 26160155 Free PMC article.
-
Potential for Kappa-Opioid Receptor Agonists to Engineer Nonaddictive Analgesics: A Narrative Review.Anesth Analg. 2021 Feb 1;132(2):406-419. doi: 10.1213/ANE.0000000000005309. Anesth Analg. 2021. PMID: 33332902 Free PMC article. Review.
References
-
- Peterson A.B., Gladden R.M., Delcher C., Spies E., Garcia-Williams A., Wang Y., Halpin J., Zibbell J., McCarty C.L., DeFiore-Hyrmer J., et al. Increases in Fentanyl-Related Overdose Deaths—Florida and Ohio, 2013–2015. MMWR Morb. Mortal. Wkly. Rep. 2016;65:844–849. doi: 10.15585/mmwr.mm6533a3. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

