Non-Glycosylated SARS-CoV-2 Omicron BA.5 Receptor Binding Domain (RBD) with a Native-like Conformation Induces a Robust Immune Response with Potent Neutralization in a Mouse Model
- PMID: 38893549
- PMCID: PMC11173568
- DOI: 10.3390/molecules29112676
Non-Glycosylated SARS-CoV-2 Omicron BA.5 Receptor Binding Domain (RBD) with a Native-like Conformation Induces a Robust Immune Response with Potent Neutralization in a Mouse Model
Abstract
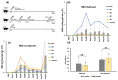
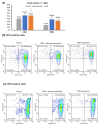
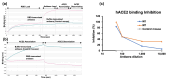
The Omicron BA.5 variant of SARS-CoV-2 is known for its high transmissibility and its capacity to evade immunity provided by vaccine protection against the (original) Wuhan strain. In our prior research, we successfully produced the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein in an E. coli expression system. Extensive biophysical characterization indicated that, even without glycosylation, the RBD maintained native-like conformational and biophysical properties. The current study explores the immunogenicity and neutralization capacity of the E. coli-expressed Omicron BA.5 RBD using a mouse model. Administration of three doses of the RBD without any adjuvant elicited high titer antisera of up to 7.3 × 105 and up to 1.6 × 106 after a booster shot. Immunization with RBD notably enhanced the population of CD44+CD62L+ T cells, indicating the generation of T cell memory. The in vitro assays demonstrated the antisera's protective efficacy through significant inhibition of the interaction between SARS-CoV-2 and its human receptor, ACE2, and through potent neutralization of a pseudovirus. These findings underscore the potential of our E. coli-expressed RBD as a viable vaccine candidate against the Omicron variant of SARS-CoV-2.
Keywords: E. coli-expression system; Omicron SARS-CoV-2; disulfide bonds; immunogenicity; limited proteolysis; molten globule state; native-like state; neutralization; non-glycosylation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., et al. Antibody Escape of SARS-CoV-2 Omicron BA.4 and BA.5 from Vaccine and BA.1 Serum. Cell. 2022;185:2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. - DOI - PMC - PubMed
-
- Servellita V., Syed A.M., Morris M.K., Brazer N., Saldhi P., Garcia-Knight M., Sreekumar B., Khalid M.M., Ciling A., Chen P.Y., et al. Neutralizing Immunity in Vaccine Breakthrough Infections from the SARS-CoV-2 Omicron and Delta Variants. Cell. 2022;185:1539. doi: 10.1016/j.cell.2022.03.019. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

