SIRPG promotes lung squamous cell carcinoma pathogenesis via M1 macrophages: a multi-omics study integrating data and Mendelian randomization
- PMID: 38894865
- PMCID: PMC11183323
- DOI: 10.3389/fonc.2024.1392417
SIRPG promotes lung squamous cell carcinoma pathogenesis via M1 macrophages: a multi-omics study integrating data and Mendelian randomization
Abstract
Background: Squamous cell carcinoma of the lung (LUSC) is a severe and highly lethal malignant tumor of the respiratory system, and its molecular mechanisms at the molecular level remain unc\lear.
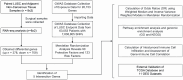
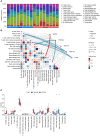
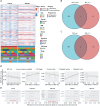
Methods: We acquired RNA-seq data from 8 surgical samples obtained from early-stage LUSC and adjacent non-cancerous tissues from 3 different centers. Utilizing Deseq2, we identified 1088 differentially expressed genes with |LogFC| > 1 and a p-value < 0.05 threshold. Furthermore, through MR analysis of Exposure Data for 26,153 Genes and 63,053 LUSC Patients, incorporating 7,838,805 SNPs as endpoints, we identified 213 genes as potential exposure factors.
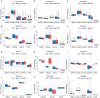
Results: After intersecting the results, we identified 5 differentially expressed genes, including GYPE, PODXL2, RNF182, SIRPG, and WNT7A. PODXL2 (OR 95% CI, 1.169 (1.040 to 1.313)) was identified as an exposed risk factor, with p-values less than 0.01 under the inverse variance weighted model. GO and KEGG analyses revealed enhanced ubiquitin-protein transferase activity and activation of pathways such as the mTOR signaling pathway and Wnt signaling pathway. Immune infiltration analysis showed downregulation of Plasma cells, T cells regulatory (Tregs), and Dendritic cells activated by the identified gene set, while an enhancement was observed in Macrophages M1. Furthermore, we externally validated the expression levels of these five genes using RNA-seq data from TCGA database and 11 GEO datasets of LUSC, and the results showed SIRPG could induce LUSC.
Conclusion: SIRPG emerged as a noteworthy exposure risk factor for LUSC. Immune infiltration analysis highlighted Macrophages M1 and mTOR signaling pathway play an important role in LUSC.
Keywords: Mendelian randomization; RNA-seq; SIRPG; immune infiltration; squamous cell carcinoma of the lung.
Copyright © 2024 Mao, Li, Wang, Yu, Han, Xiang, Zhang, Zeng, Jiang and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

