This is a preprint.
Persistent Neurological Deficits in Mouse PASC Reveal Antiviral Drug Limitations
- PMID: 38895239
- PMCID: PMC11185538
- DOI: 10.1101/2024.06.02.596989
Persistent Neurological Deficits in Mouse PASC Reveal Antiviral Drug Limitations
Abstract
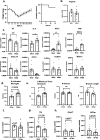
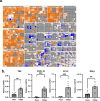
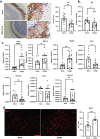
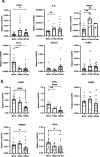
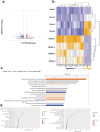
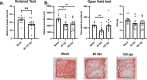
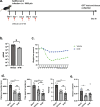
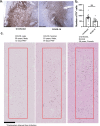
Post-Acute Sequelae of COVID-19 (PASC) encompasses persistent neurological symptoms, including olfactory and autonomic dysfunction. Here, we report chronic neurological dysfunction in mice infected with a virulent mouse-adapted SARS-CoV-2 that does not infect the brain. Long after recovery from nasal infection, we observed loss of tyrosine hydroxylase (TH) expression in olfactory bulb glomeruli and neurotransmitter levels in the substantia nigra (SN) persisted. Vulnerability of dopaminergic neurons in these brain areas was accompanied by increased levels of proinflammatory cytokines and neurobehavioral changes. RNAseq analysis unveiled persistent microglia activation, as found in human neurodegenerative diseases. Early treatment with antivirals (nirmatrelvir and molnupiravir) reduced virus titers and lung inflammation but failed to prevent neurological abnormalities, as observed in patients. Together these results show that chronic deficiencies in neuronal function in SARS-CoV-2-infected mice are not directly linked to ongoing olfactory epithelium dysfunction. Rather, they bear similarity with neurodegenerative disease, the vulnerability of which is exacerbated by chronic inflammation.
Keywords: Anosmia; Brain; Inflammation; Microglia; Neurodegeneration; Olfactory Bulb; SARS-CoV-2; Substantia Nigra; Tyrosine Hydroxylase.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest directly related to this study. MWA is a cofounder and owns shares in Aromha, Inc. He has received in kind contributions from Eli Lilly and research support from TLL Pharma. He is an SAB member of Sudo Therapeutics, and consults for BMS and Transposon.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
