This is a preprint.
Multimodal framework to resolve variants of uncertain significance in TSC2
- PMID: 38895336
- PMCID: PMC11185720
- DOI: 10.1101/2024.06.07.597916
Multimodal framework to resolve variants of uncertain significance in TSC2
Abstract
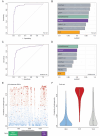
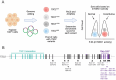
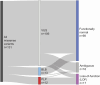
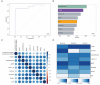
Efforts to resolve the functional impact of variants of uncertain significance (VUS) have lagged behind the identification of new VUS; as such, there is a critical need for scalable VUS resolution technologies. Computational variant effect predictors (VEPs), once trained, can predict pathogenicity for all missense variants in a gene, set of genes, or the exome. Existing tools have employed information on known pathogenic and benign variants throughout the genome to predict pathogenicity of VUS. We hypothesize that taking a gene-specific approach will improve pathogenicity prediction over globally-trained VEPs. We tested this hypothesis using the gene TSC2, whose loss of function results in tuberous sclerosis, a multisystem mTORopathy affecting about 1 in 6,000 individuals born in the United States. TSC2 has been identified as a high-priority target for VUS resolution, with (1) well-characterized molecular and patient phenotypes associated with loss-of-function variants, and (2) more than 2,700 VUS already documented in ClinVar. We developed Tuberous sclerosis classifier to Resolve variants of Uncertain Significance in T SC2 (TRUST), a machine learning model to predict pathogenicity of TSC2 missense VUS. To test whether these predictions are accurate, we further introduce curated loci prime editing (cliPE) as an accessible strategy for performing scalable multiplexed assays of variant effect (MAVEs). Using cliPE, we tested the effects of more than 200 TSC2 variants, including 106 VUS. It is highly likely this functional data alone would be sufficient to reclassify 92 VUS with most being reclassified as likely benign. We found that TRUST's classifications were correlated with the functional data, providing additional validation for the in silico predictions. We provide our pathogenicity predictions and MAVE data to aid with VUS resolution. In the near future, we plan to host these data on a public website and deposit into relevant databases such as MAVEdb as a community resource. Ultimately, this study provides a framework to complete variant effect maps of TSC1 and TSC2 and adapt this approach to other mTORopathy genes.
Figures

Similar articles
-
Using multiplexed functional data to reduce variant classification inequities in underrepresented populations.Genome Med. 2024 Dec 3;16(1):143. doi: 10.1186/s13073-024-01392-7. Genome Med. 2024. PMID: 39627863 Free PMC article.
-
Comprehensive evaluation of AlphaMissense predictions by evidence quantification for variants of uncertain significance.Front Genet. 2024 Dec 10;15:1487608. doi: 10.3389/fgene.2024.1487608. eCollection 2024. Front Genet. 2024. PMID: 39720176 Free PMC article.
-
An overview of actionable and potentially actionable TSC1 and TSC2 germline variants in an online Database.Genet Mol Biol. 2024 Feb 19;46(3 Suppl 1):e20230132. doi: 10.1590/1678-4685-GMB-2023-0132. eCollection 2024. Genet Mol Biol. 2024. PMID: 38373162 Free PMC article.
-
Navigating variants of uncertain significance in genetic dyslipidemia: how to assess and counsel patients.Curr Opin Lipidol. 2025 Apr 1;36(2):49-54. doi: 10.1097/MOL.0000000000000971. Epub 2024 Dec 18. Curr Opin Lipidol. 2025. PMID: 39950242 Review.
-
Germline Missense Variants in BRCA1: New Trends and Challenges for Clinical Annotation.Cancers (Basel). 2019 Apr 12;11(4):522. doi: 10.3390/cancers11040522. Cancers (Basel). 2019. PMID: 31013702 Free PMC article. Review.
References
-
- Weckhuysen S, Marsan E, Lambrecq V, Marchal C, Morin-Brureau M, An-Gourfinkel I, Baulac M, Fohlen M, Kallay Zetchi C, Seeck M, de la Grange P, Dermaut B, Meurs A, Thomas P, Chassoux F, Leguern E, Picard F, Baulac S. Involvement of GATOR complex genes in familial focal epilepsies and focal cortical dysplasia. Epilepsia. 2016;57(6):994–1003. Epub 20160513. doi: 10.1111/epi.13391. - DOI - PubMed
-
- Scheffer IE, Heron SE, Regan BM, Mandelstam S, Crompton DE, Hodgson BL, Licchetta L, Provini F, Bisulli F, Vadlamudi L, Gecz J, Connelly A, Tinuper P, Ricos MG, Berkovic SF, Dibbens LM. Mutations in mammalian target of rapamycin regulator DEPDC5 cause focal epilepsy with brain malformations. Ann Neurol. 2014;75(5):782–7. Epub 20140414. doi: 10.1002/ana.24126. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
