This is a preprint.
TRIM32 inhibits Venezuelan Equine Encephalitis Virus Infection by targeting a late step in viral entry
- PMID: 38895352
- PMCID: PMC11185716
- DOI: 10.1101/2024.06.04.597282
TRIM32 inhibits Venezuelan Equine Encephalitis Virus Infection by targeting a late step in viral entry
Update in
-
TRIM32 inhibits Venezuelan equine encephalitis virus infection by targeting a late step in viral entry.PLoS Pathog. 2024 Nov 11;20(11):e1012312. doi: 10.1371/journal.ppat.1012312. eCollection 2024 Nov. PLoS Pathog. 2024. PMID: 39527628 Free PMC article.
Abstract
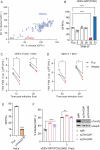
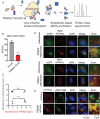
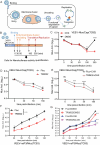
Alphaviruses are mosquito borne RNA viruses that are a reemerging public health threat. Alphaviruses have a broad host range, and can cause diverse disease outcomes like arthritis, and encephalitis. The host ubiquitin proteasome system (UPS) plays critical roles in regulating cellular processes to control the infections with various viruses, including alphaviruses. Previous studies suggest alphaviruses hijack UPS for virus infection, but the molecular mechanisms remain poorly characterized. In addition, whether certain E3 ubiquitin ligases or deubiquitinases act as alphavirus restriction factors remains poorly understood. Here, we employed a cDNA expression screen to identify E3 ubiquitin ligase TRIM32 as a novel intrinsic restriction factor against alphavirus infection, including VEEV-TC83, SINV, and ONNV. Ectopic expression of TRIM32 reduces alphavirus infection, whereas depletion of TRIM32 with CRISPR-Cas9 increases infection. We demonstrate that TRIM32 inhibits alphaviruses through a mechanism that is independent of the TRIM32-STING-IFN axis. Combining reverse genetics and biochemical assays, we found that TRIM32 interferes with genome translation after membrane fusion, prior to replication of the incoming viral genome. Furthermore, our data indicate that the monoubiquitination of TRIM32 is important for its antiviral activity. Notably, we also show two TRIM32 pathogenic mutants R394H and D487N, related to Limb-girdle muscular dystrophy (LGMD), have a loss of antiviral activity against VEEV-TC83. Collectively, these results reveal that TRIM32 acts as a novel intrinsic restriction factor suppressing alphavirus infection and provides insights into the interaction between alphaviruses and the host UPS.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
