This is a preprint.
Whole-genome DNA methylomes of Tree shrew brains reveal conserved and divergent roles of DNA methylation on sex chromosome regulation
- PMID: 38895372
- PMCID: PMC11185668
- DOI: 10.1101/2024.06.05.597676
Whole-genome DNA methylomes of Tree shrew brains reveal conserved and divergent roles of DNA methylation on sex chromosome regulation
Update in
-
Whole-genome DNA methylomes of tree shrew brains reveal conserved and divergent roles of DNA methylation on sex chromosome regulation.BMC Biol. 2024 Nov 28;22(1):277. doi: 10.1186/s12915-024-02071-0. BMC Biol. 2024. PMID: 39609804 Free PMC article.
Abstract
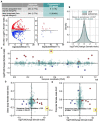
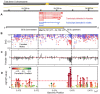
The tree shrew (Tupaia belangeri) is a promising emerging model organism in biomedical studies, notably due to their evolutionary proximity to primates. To enhance our understanding of how DNA methylation is implicated in regulation of gene expression and the X chromosome inactivation (XCI) in tree shrew brains, here we present their first genome-wide, single-base-resolution methylomes integrated with transcriptomes from prefrontal cortices. We discovered both divergent and conserved features of tree shrew DNA methylation compared to that of other mammals. DNA methylation levels of promoter and gene body regions are negatively correlated with gene expression, consistent with patterns in other mammalian brains studied. Comparing DNA methylation patterns of the female and male X chromosomes, we observed a clear and significant global reduction (hypomethylation) of DNA methylation across the entire X chromosome in females. Our data suggests that the female X hypomethylation does not directly contribute to the gene silencing of the inactivated X chromosome nor does it significantly drive sex-specific gene expression of tree shrews. However, we identified a putative regulatory region in the 5' end of the X inactive specific transcript (Xist) gene, a key gene for XCI, whose pattern of differential DNA methylation strongly relate to its differential expression between male and female tree shrews. We show that differential methylation of this region is conserved across different species. Moreover, we provide evidence suggesting that the observed difference between human and tree shrew X-linked promoter methylation is associated with the difference in genomic CpG contents. Our study offers novel information on genomic DNA methylation of tree shrews, as well as insights into the evolution of X chromosome regulation in mammals.
Keywords: DNA methylation; X chromosome; gene expression; tree shrew (Tupaia belangeri).
Figures



References
-
- Al Adhami H., Bardet A. F., Dumas M., Cleroux E., Guibert S., Fauque P., Acloque H., & Weber M. (2022). A comparative methylome analysis reveals conservation and divergence of DNA methylation patterns and functions in vertebrates. BMC Biology, 20(1), 70. 10.1186/s12915-022-01270-x - DOI - PMC - PubMed
-
- Brown C. J., Hendrich B. D., Rupert J. L., Lafrenière R. G., Xing Y., Lawrence J., & Willard H. F. (1992). The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell, 71(3), 527–542. 10.1016/0092-8674(92)90520-m - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
