This is a preprint.
BMAL1-HIF2α heterodimers contribute to ccRCC
- PMID: 38895384
- PMCID: PMC11185709
- DOI: 10.1101/2024.06.07.597806
BMAL1-HIF2α heterodimers contribute to ccRCC
Abstract
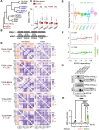
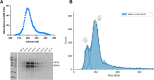
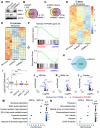
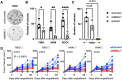
Circadian disruption enhances cancer risk, and many tumors exhibit disordered circadian gene expression. We show rhythmic gene expression is unexpectedly robust in clear cell renal cell carcinoma (ccRCC). Furthermore, the clock gene BMAL1 is higher in ccRCC than in healthy kidneys, unlike in other tumor types. BMAL1 is closely related to ARNT, and we show that BMAL1-HIF2α regulates a subset of HIF2α target genes in ccRCC cells. Depletion of BMAL1 reprograms HIF2α chromatin association and target gene expression and reduces ccRCC growth in culture and in xenografts. Analysis of pre-existing data reveals higher BMAL1 in patient-derived xenografts that are sensitive to growth suppression by a HIF2α antagonist (PT2399). We show that BMAL1-HIF2α is more sensitive than ARNT-HIF2α to suppression by PT2399, and increasing BMAL1 sensitizes 786O cells to growth inhibition by PT2399. Together, these findings indicate that an alternate HIF2α heterodimer containing the circadian partner BMAL1 contributes to HIF2α activity, growth, and sensitivity to HIF2α antagonist drugs in ccRCC cells.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures


Similar articles
-
BMAL1-HIF2α heterodimers contribute to ccRCC.Res Sq [Preprint]. 2024 Jul 16:rs.3.rs-4651047. doi: 10.21203/rs.3.rs-4651047/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Jul 1;16(1):5834. doi: 10.1038/s41467-025-60904-0. PMID: 39070610 Free PMC article. Updated. Preprint.
-
BMAL1 and ARNT enable circadian HIF2α responses in clear cell renal cell carcinoma.Nat Commun. 2025 Jul 1;16(1):5834. doi: 10.1038/s41467-025-60904-0. Nat Commun. 2025. PMID: 40595592 Free PMC article.
-
Aberrant activation of m6A demethylase FTO renders HIF2αlow/- clear cell renal cell carcinoma sensitive to BRD9 inhibitors.Sci Transl Med. 2021 Sep 29;13(613):eabf6045. doi: 10.1126/scitranslmed.abf6045. Epub 2021 Sep 29. Sci Transl Med. 2021. PMID: 34586831
-
The role of HIF1α in renal cell carcinoma tumorigenesis.J Mol Med (Berl). 2014 Aug;92(8):825-36. doi: 10.1007/s00109-014-1180-z. Epub 2014 Jun 12. J Mol Med (Berl). 2014. PMID: 24916472 Free PMC article. Review.
-
Regulatory Effects of Clock and Bmal1 on Circadian Rhythmic TLR Expression.Int Rev Immunol. 2023;42(2):101-112. doi: 10.1080/08830185.2021.1931170. Epub 2021 Sep 20. Int Rev Immunol. 2023. PMID: 34544330 Review.
References
-
- Dong Z. et al. Targeting Glioblastoma Stem Cells through Disruption of the Circadian Clock. Cancer Discov 9, 1556–1573 (2019). 10.1158/2159-8290.CD-19-0215 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
