This is a preprint.
Mechanism of how the universal module XMAP215 γ-TuRC nucleates microtubules
- PMID: 38895418
- PMCID: PMC11185565
- DOI: 10.1101/2024.06.03.597159
Mechanism of how the universal module XMAP215 γ-TuRC nucleates microtubules
Abstract
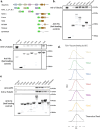
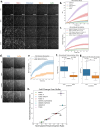
It has become increasingly evident in recent years that nucleation of microtubules from a diverse set of MTOCs requires both the γ-tubulin ring complex (γ-TuRC) and the microtubule polymerase XMAP215. Despite their essentiality, little is known about how these nucleation factors interact and work together to generate microtubules. Using biochemical domain analysis of XMAP215 and structural approaches, we find that a sixth TOG domain in XMAP215 binds γ-TuRC via γ-tubulin as part of a broader interaction involving the C-terminal region. Moreover, TOG6 is required for XMAP215 to promote nucleation from γ-TuRC to its full extent. Interestingly, we find that XMAP215 also depends strongly on TOG5 for microtubule lattice binding and nucleation. Accordingly, we report a cryo-EM structure of TOG5 bound to the microtubule lattice that reveals promotion of lateral interactions between tubulin dimers. Finally, we find that while XMAP215 constructs' effects on nucleation are generally proportional to their effects on polymerization, formation of a direct complex with γ-TuRC allows cooperative nucleation activity. Thus, we propose that XMAP215's C-terminal TOGs 5 and 6 play key roles in promoting nucleation by promoting formation of longitudinal and lateral bonds in γ-TuRC templated nascent microtubules at cellular MTOCs.
Figures




Similar articles
-
XMAP215 is a microtubule nucleation factor that functions synergistically with the γ-tubulin ring complex.Nat Cell Biol. 2018 May;20(5):575-585. doi: 10.1038/s41556-018-0091-6. Epub 2018 Apr 25. Nat Cell Biol. 2018. PMID: 29695792 Free PMC article.
-
The transition state and regulation of γ-TuRC-mediated microtubule nucleation revealed by single molecule microscopy.Elife. 2020 Jun 15;9:e54253. doi: 10.7554/eLife.54253. Elife. 2020. PMID: 32538784 Free PMC article.
-
Microtubule nucleation: The waltz between γ-tubulin ring complex and associated proteins.Curr Opin Cell Biol. 2021 Feb;68:124-131. doi: 10.1016/j.ceb.2020.10.004. Epub 2020 Nov 12. Curr Opin Cell Biol. 2021. PMID: 33190097 Review.
-
Structure of the microtubule anchoring factor NEDD1 bound to the γ-tubulin ring complex.bioRxiv [Preprint]. 2024 Nov 5:2024.11.05.622067. doi: 10.1101/2024.11.05.622067. bioRxiv. 2024. Update in: J Cell Biol. 2025 Aug 4;224(8):e202410206. doi: 10.1083/jcb.202410206. PMID: 39574704 Free PMC article. Updated. Preprint.
-
Molecular insight into how γ-TuRC makes microtubules.J Cell Sci. 2021 Jul 15;134(14):jcs245464. doi: 10.1242/jcs.245464. Epub 2021 Jul 23. J Cell Sci. 2021. PMID: 34297125 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
