This is a preprint.
Cross-species modeling of plant genomes at single nucleotide resolution using a pre-trained DNA language model
- PMID: 38895432
- PMCID: PMC11185591
- DOI: 10.1101/2024.06.04.596709
Cross-species modeling of plant genomes at single nucleotide resolution using a pre-trained DNA language model
Update in
-
Cross-species modeling of plant genomes at single-nucleotide resolution using a pretrained DNA language model.Proc Natl Acad Sci U S A. 2025 Jun 17;122(24):e2421738122. doi: 10.1073/pnas.2421738122. Epub 2025 Jun 9. Proc Natl Acad Sci U S A. 2025. PMID: 40489624
Abstract
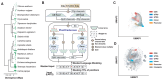
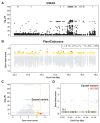
Interpreting function and fitness effects in diverse plant genomes requires transferable models. Language models (LMs) pre-trained on large-scale biological sequences can learn evolutionary conservation and offer cross-species prediction better than supervised models through fine-tuning limited labeled data. We introduce PlantCaduceus, a plant DNA LM based on the Caduceus and Mamba architectures, pre-trained on a curated dataset of 16 Angiosperm genomes. Fine-tuning PlantCaduceus on limited labeled Arabidopsis data for four tasks, including predicting translation initiation/termination sites and splice donor and acceptor sites, demonstrated high transferability to 160 million year diverged maize, outperforming the best existing DNA LM by 1.45 to 7.23-fold. PlantCaduceus is competitive to state-of-the-art protein LMs in terms of deleterious mutation identification, and is threefold better than PhyloP. Additionally, PlantCaduceus successfully identifies well-known causal variants in both Arabidopsis and maize. Overall, PlantCaduceus is a versatile DNA LM that can accelerate plant genomics and crop breeding applications.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures



References
-
- Sun Y., Shang L., Zhu Q.-H., Fan L. & Guo L. Twenty years of plant genome sequencing: achievements and challenges. Trends Plant Sci. 27, 391–401 (2022). - PubMed
-
- Soltis P. S. & Soltis D. E. Plant genomes: Markers of evolutionary history and drivers of evolutionary change. Plants People Planet 3, 74–82 (2021).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
