This is a preprint.
Structural polymorphism and diversity of human segmental duplications
- PMID: 38895457
- PMCID: PMC11185583
- DOI: 10.1101/2024.06.04.597452
Structural polymorphism and diversity of human segmental duplications
Update in
-
Structural polymorphism and diversity of human segmental duplications.Nat Genet. 2025 Feb;57(2):390-401. doi: 10.1038/s41588-024-02051-8. Epub 2025 Jan 8. Nat Genet. 2025. PMID: 39779957 Free PMC article.
Abstract
Segmental duplications (SDs) contribute significantly to human disease, evolution, and diversity yet have been difficult to resolve at the sequence level. We present a population genetics survey of SDs by analyzing 170 human genome assemblies where the majority of SDs are fully resolved using long-read sequence assembly. Excluding the acrocentric short arms, we identify 173.2 Mbp of duplicated sequence (47.4 Mbp not present in the telomere-to-telomere reference) distinguishing fixed from structurally polymorphic events. We find that intrachromosomal SDs are among the most variable with rare events mapping near their progenitor sequences. African genomes harbor significantly more intrachromosomal SDs and are more likely to have recently duplicated gene families with higher copy number when compared to non-African samples. A comparison to a resource of 563 million full-length Iso-Seq reads identifies 201 novel, potentially protein-coding genes corresponding to these copy number polymorphic SDs.
Conflict of interest statement
COMPETING INTERESTS E.E.E. is a scientific advisory board (SAB) member of Variant Bio, Inc. C.L. is an SAB member of Nabsys and Genome Insight. The other authors declare no competing interests.
Figures
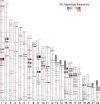
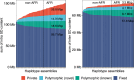
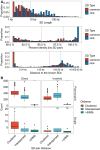


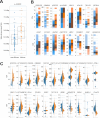

References
-
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). - PubMed
-
- Eichler E. Interchromosomal duplications of the adrenoleukodystrophy locus: a phenomenon of pericentromeric plasticity. Hum. Mol. Genet. 6, 991–1002 (1997). - PubMed
-
- Trask B. J. et al. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum. Mol. Genet. 7, 13–26 (1998). - PubMed
-
- Church D. M. A next-generation human genome sequence. Science 376, 34–35 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
