Tubulin-binding region alters tau-lipid interactions and changes toxicity of tau fibrils formed in the presence of phosphatidylserine lipids
- PMID: 38895991
- PMCID: PMC11187861
- DOI: 10.1002/pro.5078
Tubulin-binding region alters tau-lipid interactions and changes toxicity of tau fibrils formed in the presence of phosphatidylserine lipids
Abstract
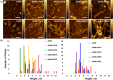
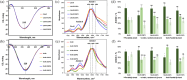
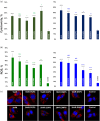
Alzheimer's disease is the fastest-growing neurodegenerative disease that affects over six million Americans. The abnormal aggregation of amyloid β peptide and Tau protein is the expected molecular cause of the loss of neurons in brains of AD patients. A growing body of evidence indicates that lipids can alter the aggregation rate of amyloid β peptide and modify the toxicity of amyloid β aggregates. However, the role of lipids in Tau aggregation remains unclear. In this study, we utilized a set of biophysical methods to determine the extent to which phospatidylserine (PS) altered the aggregation properties of Tau isoforms with one (1N4R) and two (2N4R) N terminal inserts that enhance the binding of Tau to tubulin. We found that the length and saturation of fatty acids (FAs) in PS altered the aggregation rate of 2N4R isoform, while no changes in the aggregation rate of 1N4R were observed. These results indicate that N terminal inserts play an important role in protein-lipid interactions. We also found that PS could change the toxicity of 1N4R and 2N4R Tau fibrils, as well as alter molecular mechanisms by which these aggregates exert cytotoxicity to neurons. Finally, we found that although Tau fibrils formed in the presence and absence of PS endocytosed by cells, only fibril species that were formed in the presence of PS exert strong impairment of the cell mitochondria.
Keywords: 1N4R tau; 2N4R tau; AFM‐IR; fibrils; oligomers; phosphatidylserine; toxicity.
© 2024 The Protein Society.
Figures



Similar articles
-
Length and saturation of fatty acids in phosphatidylserine determine the rate of lysozyme aggregation simultaneously altering the structure and toxicity of amyloid oligomers and fibrils.Protein Sci. 2023 Aug;32(8):e4717. doi: 10.1002/pro.4717. Protein Sci. 2023. PMID: 37402649 Free PMC article.
-
Under Heparin-Free Conditions Unsaturated Phospholipids Inhibit the Aggregation of 1N4R and 2N4R Tau.J Phys Chem Lett. 2024 Aug 22;15(33):8577-8583. doi: 10.1021/acs.jpclett.4c01718. Epub 2024 Aug 14. J Phys Chem Lett. 2024. PMID: 39140785 Free PMC article.
-
Discovery of small molecule benzothiazole and indole derivatives tackling tau 2N4R and α-synuclein fibrils.Bioorg Med Chem. 2024 Feb 15;100:117613. doi: 10.1016/j.bmc.2024.117613. Epub 2024 Jan 28. Bioorg Med Chem. 2024. PMID: 38330847 Free PMC article.
-
Heat shock proteins regulates Tau protein aggregation in Alzheimer's disease.Adv Protein Chem Struct Biol. 2025;146:161-178. doi: 10.1016/bs.apcsb.2024.08.003. Epub 2024 Sep 11. Adv Protein Chem Struct Biol. 2025. PMID: 40610073 Review.
-
Protein aggregation and neurodegenerative disease: Structural outlook for the novel therapeutics.Proteins. 2025 Aug;93(8):1314-1329. doi: 10.1002/prot.26561. Epub 2023 Aug 2. Proteins. 2025. PMID: 37530227 Free PMC article. Review.
Cited by
-
Tubulin-Binding Region Modulates Cholesterol-Triggered Aggregation of Tau Proteins.J Neurochem. 2025 Jan;169(1):e16294. doi: 10.1111/jnc.16294. J Neurochem. 2025. PMID: 39777699
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

