Interplay between protease and reverse transcriptase dimerization in a model HIV-1 polyprotein
- PMID: 38896002
- PMCID: PMC11187873
- DOI: 10.1002/pro.5080
Interplay between protease and reverse transcriptase dimerization in a model HIV-1 polyprotein
Abstract
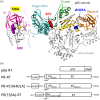
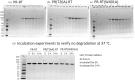
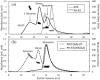
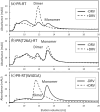
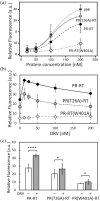
The Gag-Pol polyprotein in human immunodeficiency virus type I (HIV-1) encodes enzymes that are essential for virus replication: protease (PR), reverse transcriptase (RT), and integrase (IN). The mature forms of PR, RT and IN are homodimer, heterodimer and tetramer, respectively. The precise mechanism underlying the formation of dimer or tetramer is not yet understood. Here, to gain insight into the dimerization of PR and RT in the precursor, we prepared a model precursor, PR-RT, incorporating an inactivating mutation at the PR active site, D25A, and including two residues in the p6* region, fused to a SUMO-tag, at the N-terminus of the PR region. We also prepared two mutants of PR-RT containing a dimer dissociation mutation either in the PR region, PR(T26A)-RT, or in the RT region, PR-RT(W401A). Size exclusion chromatography showed both monomer and dimer fractions in PR-RT and PR(T26A)-RT, but only monomer in PR-RT(W401A). SEC experiments of PR-RT in the presence of protease inhibitor, darunavir, significantly enhanced the dimerization. Additionally, SEC results suggest an estimated PR-RT dimer dissociation constant that is higher than that of the mature RT heterodimer, p66/p51, but slightly lower than the premature RT homodimer, p66/p66. Reverse transcriptase assays and RT maturation assays were performed as tools to assess the effects of the PR dimer-interface on these functions. Our results consistently indicate that the RT dimer-interface plays a crucial role in the dimerization in PR-RT, whereas the PR dimer-interface has a lesser role.
Keywords: Gag‐Pol polyprotein; HIV‐1; darunavir; dimerization; inhibitor; protease; reverse transcriptase.
© 2024 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bavand MR, Wagner R, Richmond TJ. HIV‐1 reverse transcriptase: polymerization properties of the p51 homodimer compared to the p66/p51 heterodimer. Biochemistry. 1993;32:10543–10552. - PubMed
-
- Bhavyasri KSM, Sumakanth M. Simultaneous method development, validation and stress studies of darunavir and ritonavir in bulk and combined dosage form using UV spectroscopy. Scholars Academic Journal of Pharmacy. 2020;9:244–252.
-
- Chattopadhyay D, Evans DB, Deibel MR, Vosters AF, Eckenrode FM, Einspahr HM, et al. Purification and characterization of heterodimeric human immunodeficiency virus type 1 (HIV‐1) reverse transcriptase produced by in vitro processing of p66 with recombinant HIV‐1 protease. Journal of Biological Chemistry. 1992;267:14227–14232. - PubMed
-
- Cherepanov P, Maertens G, Proost P, Devreese B, van Beeumen J, Engelborghs Y, et al. HIV‐1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. Journal of Biological Chemistry. 2003;278:372–381. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

