Machine learning of dissection photographs and surface scanning for quantitative 3D neuropathology
- PMID: 38896568
- PMCID: PMC11186625
- DOI: 10.7554/eLife.91398
Machine learning of dissection photographs and surface scanning for quantitative 3D neuropathology
Abstract
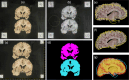
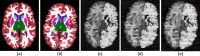
We present open-source tools for three-dimensional (3D) analysis of photographs of dissected slices of human brains, which are routinely acquired in brain banks but seldom used for quantitative analysis. Our tools can: (1) 3D reconstruct a volume from the photographs and, optionally, a surface scan; and (2) produce a high-resolution 3D segmentation into 11 brain regions per hemisphere (22 in total), independently of the slice thickness. Our tools can be used as a substitute for ex vivo magnetic resonance imaging (MRI), which requires access to an MRI scanner, ex vivo scanning expertise, and considerable financial resources. We tested our tools on synthetic and real data from two NIH Alzheimer's Disease Research Centers. The results show that our methodology yields accurate 3D reconstructions, segmentations, and volumetric measurements that are highly correlated to those from MRI. Our method also detects expected differences between post mortem confirmed Alzheimer's disease cases and controls. The tools are available in our widespread neuroimaging suite 'FreeSurfer' (https://surfer.nmr.mgh.harvard.edu/fswiki/PhotoTools).
Keywords: dissection photography; human; machine learning; neuroscience; surface scanning; volumetry.
Plain language summary
Every year, thousands of human brains are donated to science. These brains are used to study normal aging, as well as neurological diseases like Alzheimer’s or Parkinson’s. Donated brains usually go to ‘brain banks’, institutions where the brains are dissected to extract tissues relevant to different diseases. During this process, it is routine to take photographs of brain slices for archiving purposes. Often, studies of dead brains rely on qualitative observations, such as ‘the hippocampus displays some atrophy’, rather than concrete ‘numerical’ measurements. This is because the gold standard to take three-dimensional measurements of the brain is magnetic resonance imaging (MRI), which is an expensive technique that requires high expertise – especially with dead brains. The lack of quantitative data means it is not always straightforward to study certain conditions. To bridge this gap, Gazula et al. have developed an openly available software that can build three-dimensional reconstructions of dead brains based on photographs of brain slices. The software can also use machine learning methods to automatically extract different brain regions from the three-dimensional reconstructions and measure their size. These data can be used to take precise quantitative measurements that can be used to better describe how different conditions lead to changes in the brain, such as atrophy (reduced volume of one or more brain regions). The researchers assessed the accuracy of the method in two ways. First, they digitally sliced MRI-scanned brains and used the software to compute the sizes of different structures based on these synthetic data, comparing the results to the known sizes. Second, they used brains for which both MRI data and dissection photographs existed and compared the measurements taken by the software to the measurements obtained with MRI images. Gazula et al. show that, as long as the photographs satisfy some basic conditions, they can provide good estimates of the sizes of many brain structures. The tools developed by Gazula et al. are publicly available as part of FreeSurfer, a widespread neuroimaging software that can be used by any researcher working at a brain bank. This will allow brain banks to obtain accurate measurements of dead brains, allowing them to cheaply perform quantitative studies of brain structures, which could lead to new findings relating to neurodegenerative diseases.
© 2023, Gazula et al.
Conflict of interest statement
HG, HT, BB, YB, JW, RH, LD, AC, EM, CL, MK, MM, ER, EB, MM, TC, DO, MF, SY, KV, AD, CM, CK, JI No competing interests declared, BF BF has a financial interest in CorticoMetrics, a company developing brain MRI measurementtechnology; his interests are reviewed and managed by Massachusetts General Hospital, BH Reviewing editor, eLife
Figures



Update of
-
Machine learning of dissection photographs and surface scanning for quantitative 3D neuropathology.bioRxiv [Preprint]. 2024 Jan 30:2023.06.08.544050. doi: 10.1101/2023.06.08.544050. bioRxiv. 2024. Update in: Elife. 2024 Jun 19;12:RP91398. doi: 10.7554/eLife.91398. PMID: 37333251 Free PMC article. Updated. Preprint.
References
-
- Abadi M, Barham P, Chen J, Chen Z, Davis A. Tensorflow: A system for large-scale machine learning. Symposium on Operating Systems Design and Implementation; 2016. pp. 265–283.
-
- Billot B, Greve D, Van Leemput K, Fischl B, Iglesias JE, Dalca A. A Learning Strategy for Contrast-agnostic MRI Segmentation. Medical Imaging with Deep Learning; 2020. pp. 75–93.
MeSH terms
Grants and funding
- U01MH117023/NH/NIH HHS/United States
- R01EB031114/NH/NIH HHS/United States
- R01NS083534/NH/NIH HHS/United States
- R01 MH121885/NH/NIH HHS/United States
- R01 MH123195/NH/NIH HHS/United States
- R01 EB023281/EB/NIBIB NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- R01EB019956/NH/NIH HHS/United States
- Grant Ref 2021UPC-MS-67573/Politècnica de Catalunya
- U01 MH093765/MH/NIMH NIH HHS/United States
- R01 NS070963/NS/NINDS NIH HHS/United States
- U01 NS086625/NS/NINDS NIH HHS/United States
- R21 EB018907/EB/NIBIB NIH HHS/United States
- ARUK-IRG2019A-003/Alzheimer's Research UK
- R21NS072652/NH/NIH HHS/United States
- R01 AG064027/AG/NIA NIH HHS/United States
- 1S10RR023043/NH/NIH HHS/United States
- R01 AG016495/AG/NIA NIH HHS/United States
- P30 AG062421/AG/NIA NIH HHS/United States
- 1R01EB023281/NH/NIH HHS/United States
- R01AG016495/NH/NIH HHS/United States
- R01 NS112161/NS/NINDS NIH HHS/United States
- S10 RR019307/RR/NCRR NIH HHS/United States
- R01NS112161/NH/NIH HHS/United States
- R01 AG070988/AG/NIA NIH HHS/United States
- P30AG066509 (UW ADRC)/NH/NIH HHS/United States
- RF1 MH123195/MH/NIMH NIH HHS/United States
- 1S10RR019307/NH/NIH HHS/United States
- R01EB006758/NH/NIH HHS/United States
- R01AG070988/AG/NIA NIH HHS/United States
- RF1 MH121885/MH/NIMH NIH HHS/United States
- 5U01NS086625/NH/NIH HHS/United States
- R01NS105820/NH/NIH HHS/United States
- R01 NS105820/NS/NINDS NIH HHS/United States
- 1R56AG064027/NH/NIH HHS/United States
- 1R01AG070988/NH/NIH HHS/United States
- K08AG065426/NH/NIH HHS/United States
- U19 AG060909/AG/NIA NIH HHS/United States
- 1RF1MH123195/NH/NIH HHS/United States
- 5R01AG008122/NH/NIH HHS/United States
- P30 AG066509/AG/NIA NIH HHS/United States
- ERC Starting Grant 677697/European Union
- P30AG062421/AG/NIA NIH HHS/United States
- U19AG066567/NH/NIH HHS/United States
- R01 EB019956/EB/NIBIB NIH HHS/United States
- R56 AG064027/AG/NIA NIH HHS/United States
- K08 AG065426/AG/NIA NIH HHS/United States
- P41EB030006/NH/NIH HHS/United States
- 5U01MH093765/NH/NIH HHS/United States
- P41 EB030006/EB/NIBIB NIH HHS/United States
- R01 EB031114/EB/NIBIB NIH HHS/United States
- 1R01AG064027/NH/NIH HHS/United States
- 5U24NS10059103/NH/NIH HHS/United States
- R01NS070963/NH/NIH HHS/United States
- U19AG060909/NH/NIH HHS/United States
- U01 MH117023/MH/NIMH NIH HHS/United States
- R21 NS072652/NS/NINDS NIH HHS/United States
- UM1MH130981/NH/NIH HHS/United States
- S10 RR023043/RR/NCRR NIH HHS/United States
- 1S10RR023401/NH/NIH HHS/United States
- U19 AG066567/AG/NIA NIH HHS/United States
- RF1MH123195/NH/NIH HHS/United States
- R21EB018907/NH/NIH HHS/United States
- R01 EB006758/EB/NIBIB NIH HHS/United States
- R01 NS083534/NS/NINDS NIH HHS/United States
- S10 RR023401/RR/NCRR NIH HHS/United States
- R01NS0525851/NH/NIH HHS/United States
- UM1 MH130981/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

