Posttranslational regulation of photosynthetic activity via the TOR kinase in plants
- PMID: 38896607
- PMCID: PMC11186500
- DOI: 10.1126/sciadv.adj3268
Posttranslational regulation of photosynthetic activity via the TOR kinase in plants
Abstract
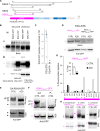
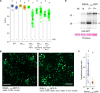
Chloroplasts are the powerhouse of the plant cell, and their activity must be matched to plant growth to avoid photooxidative damage. We have identified a posttranslational mechanism linking the eukaryotic target of rapamycin (TOR) kinase that promotes growth and the guanosine tetraphosphate (ppGpp) signaling pathway of prokaryotic origins that regulates chloroplast activity and photosynthesis in particular. We find that RelA SpoT homolog 3 (RSH3), a nuclear-encoded enzyme responsible for ppGpp biosynthesis, interacts directly with the TOR complex via a plant-specific amino-terminal region which is phosphorylated in a TOR-dependent manner. Down-regulating TOR activity causes a rapid increase in ppGpp synthesis in RSH3 overexpressors and reduces photosynthetic capacity in an RSH-dependent manner in wild-type plants. The TOR-RSH3 signaling axis therefore regulates the equilibrium between chloroplast activity and plant growth, setting a precedent for the regulation of organellar function by TOR.
Figures

References
-
- Bange G., Brodersen D. E., Liuzzi A., Steinchen W., Two P or not two P: Understanding regulation by the bacterial second messengers (p)ppGpp. Annu. Rev. Microbiol. 75, 383–406 (2021). - PubMed
-
- Maekawa M., Honoki R., Ihara Y., Sato R., Oikawa A., Kanno Y., Ohta H., Seo M., Saito K., Masuda S., Impact of the plastidial stringent response in plant growth and stress responses. Nat. Plants 1, 15167 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

