The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain
- PMID: 38896613
- PMCID: PMC11186497
- DOI: 10.1126/sciadv.adm9404
The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain
Abstract
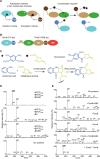
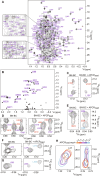
In the quest for new bioactive substances, nonribosomal peptide synthetases (NRPS) provide biodiversity by synthesizing nonproteinaceous peptides with high cellular activity. NRPS machinery consists of multiple modules, each catalyzing a unique series of chemical reactions. Incomplete understanding of the biophysical principles orchestrating these reaction arrays limits the exploitation of NRPSs in synthetic biology. Here, we use nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry to solve the conundrum of how intermodular recognition is coupled with loaded carrier protein specificity in the tomaymycin NRPS. We discover an adaptor domain that directly recruits the loaded carrier protein from the initiation module to the elongation module and reveal its mechanism of action. The adaptor domain of the type found here has specificity rules that could potentially be exploited in the design of engineered NRPS machinery.
Figures



Similar articles
-
The structure of the monobactam-producing thioesterase domain of SulM forms a unique complex with the upstream carrier protein domain.J Biol Chem. 2024 Aug;300(8):107489. doi: 10.1016/j.jbc.2024.107489. Epub 2024 Jun 20. J Biol Chem. 2024. PMID: 38908753 Free PMC article.
-
Modular catalytic activity of nonribosomal peptide synthetases depends on the dynamic interaction between adenylation and condensation domains.Structure. 2024 Apr 4;32(4):440-452.e4. doi: 10.1016/j.str.2024.01.010. Epub 2024 Feb 9. Structure. 2024. PMID: 38340732
-
Chemical Logic of Peptide Branching by Iterative Nonlinear Nonribosomal Peptide Synthetases.Biochemistry. 2025 Feb 4;64(3):719-734. doi: 10.1021/acs.biochem.4c00749. Epub 2025 Jan 23. Biochemistry. 2025. PMID: 39847710 Review.
-
Probing Substrate-Loaded Carrier Proteins by Nuclear Magnetic Resonance.Methods Mol Biol. 2023;2670:235-253. doi: 10.1007/978-1-0716-3214-7_12. Methods Mol Biol. 2023. PMID: 37184708
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
Cited by
-
NMR chemical shift assignment of the IMLV methyl groups of a di-domain of the Tomaymycin non-ribosomal peptide synthetase.Biomol NMR Assign. 2025 Jun;19(1):153-164. doi: 10.1007/s12104-025-10231-8. Epub 2025 Apr 25. Biomol NMR Assign. 2025. PMID: 40278976 Free PMC article.
-
The structural basis of substrate selectivity of the acinetobactin biosynthetic adenylation domain, BasE.J Biol Chem. 2025 Apr;301(4):108413. doi: 10.1016/j.jbc.2025.108413. Epub 2025 Mar 15. J Biol Chem. 2025. PMID: 40096888 Free PMC article.
References
-
- Süssmuth R. D., Mainz A., Nonribosomal peptide synthesis-principles and prospects. Angew. Chem. Int. Ed. Engl. 56, 3770–3821 (2017). - PubMed
-
- Brown A. S., Calcott M. J., Owen J. G., Ackerley D. F., Structural, functional and evolutionary perspectives on effective re-engineering of non-ribosomal peptide synthetase assembly lines. Nat. Prod. Rep. 35, 1210–1228 (2018). - PubMed
-
- Marahiel M. A., Stachelhaus T., Mootz H. D., Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2674 (1997). - PubMed
-
- Bloudoff K., Schmeing T. M., Structural and functional aspects of the nonribosomal peptide synthetase condensation domain superfamily: Discovery, dissection and diversity. Biochim. Biophys. Acta Proteins Proteomics 1865, 1587–1604 (2017). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

