Prolonged Cadmium Exposure and Osteoclastogenesis: A Mechanistic Mouse and in Vitro Study
- PMID: 38896780
- PMCID: PMC11218709
- DOI: 10.1289/EHP13849
Prolonged Cadmium Exposure and Osteoclastogenesis: A Mechanistic Mouse and in Vitro Study
Abstract
Background: Cadmium (Cd) is a highly toxic and widespread environmental oxidative stressor that causes a myriad of health problems, including osteoporosis and bone damage. Although nuclear factor erythroid 2-related factor 2 (NRF2) and its Cap 'n' Collar and basic region Leucine Zipper (CNC-bZIP) family member nuclear factor erythroid 2-related factor 1 (NRF1) coordinate various stress responses by regulating the transcription of a variety of antioxidant and cytoprotective genes, they play distinct roles in bone metabolism and remodeling. However, the precise roles of both transcription factors in bone loss induced by prolonged Cd exposure remain unclear.
Objectives: We aimed to understand the molecular mechanisms underlying Cd-induced bone loss, focusing mainly on the roles of NRF2 and NRF1 in osteoclastogenesis provoked by Cd.
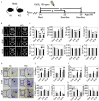
Methods: Male wild-type (WT), global Nrf2-knockout () and myeloid-specific Nrf2 knockout [Nrf2(M)-KO] mice were administered Cd (50 or ) via drinking water for 8 or 16 wk, followed by micro-computed tomography, histological analyses, and plasma biochemical testing. Osteoclastogenesis was evaluated using bone marrow-derived osteoclast progenitor cells (BM-OPCs) and RAW 264.7 cells in the presence of Cd (10 or ) with a combination of genetic and chemical modulations targeting NRF2 and NRF1.
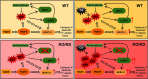
Results: Compared with relevant control mice, global or Nrf2(M)-KO mice showed exacerbated bone loss and augmented osteoclast activity following exposure to Cd in drinking water for up to 16 wk. In vitro osteoclastogenic analyses suggested that Nrf2-deficient BM-OPCs and RAW 264.7 cells responded more robustly to low levels of Cd (up to ) with regard to osteoclast differentiation compared with WT cells. Further mechanistic studies supported a compensatory up-regulation of long isoform of NRF1 (L-NRF1) and subsequent induction of nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 (NFATc1) as the key molecular events in the Nrf2 deficiency-worsened and Cd-provoked osteoclastogenesis. L-Nrf1 silenced (via lentiviral means) Nrf2-knockdown (KD) RAW cells exposed to Cd showed dramatically different NFATc1 and subsequent osteoclastogenesis outcomes compared with the cells of Nrf2-KD alone exposed to Cd, suggesting a mitigating effect of the Nrf1 silencing. In addition, suppression of reactive oxygen species by exogenous antioxidants -acetyl-l-cysteine () and mitoquinone mesylate (MitoQ; ) mitigated the L-NRF1-associated effects on NFATc1-driven osteoclastogenesis outcomes in Cd-exposed Nrf2-KD cells.
Conclusions: This in vivo and in vitro study supported the authors' hypothesis that Cd exposure caused bone loss, in which NRF2 and L-NRF1 responded to Cd and osteoclastogenic stimuli in a cooperative, but contradictive, manner to coordinate Nfatc1 expression, osteoclastogenesis and thus bone homeostasis. Our study suggests a novel strategy targeting NRF2 and L-NRF1 to prevent and treat the bone toxicity of Cd. https://doi.org/10.1289/EHP13849.
Figures




Similar articles
-
Nrf2 deficiency aggravates the increase in osteoclastogenesis and bone loss induced by inorganic arsenic.Toxicol Appl Pharmacol. 2019 Mar 15;367:62-70. doi: 10.1016/j.taap.2019.02.003. Epub 2019 Feb 8. Toxicol Appl Pharmacol. 2019. PMID: 30742845
-
Carnosic acid attenuates RANKL-induced oxidative stress and osteoclastogenesis via induction of Nrf2 and suppression of NF-κB and MAPK signalling.J Mol Med (Berl). 2017 Oct;95(10):1065-1076. doi: 10.1007/s00109-017-1553-1. Epub 2017 Jul 4. J Mol Med (Berl). 2017. PMID: 28674855
-
Dehydrocostus lactone inhibits NFATc1 via regulation of IKK, JNK, and Nrf2, thereby attenuating osteoclastogenesis.BMB Rep. 2020 Apr;53(4):218-222. doi: 10.5483/BMBRep.2020.53.4.220. BMB Rep. 2020. PMID: 31964469 Free PMC article.
-
Role of Nrf1 in antioxidant response element-mediated gene expression and beyond.Toxicol Appl Pharmacol. 2010 Apr 1;244(1):16-20. doi: 10.1016/j.taap.2009.07.034. Epub 2009 Aug 6. Toxicol Appl Pharmacol. 2010. PMID: 19665035 Free PMC article. Review.
-
Targeting Sirt1, AMPK, Nrf2, CK2, and Soluble Guanylate Cyclase with Nutraceuticals: A Practical Strategy for Preserving Bone Mass.Int J Mol Sci. 2022 Apr 26;23(9):4776. doi: 10.3390/ijms23094776. Int J Mol Sci. 2022. PMID: 35563167 Free PMC article. Review.
Cited by
-
Invited Perspective: New Insight into Cadmium-Related Osteoporosis Yields Hope for Prevention and Therapy.Environ Health Perspect. 2024 Jun;132(6):61301. doi: 10.1289/EHP15263. Epub 2024 Jun 19. Environ Health Perspect. 2024. PMID: 38896781 Free PMC article. No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

