Oncometabolite 2-hydroxyglutarate suppresses basal protein levels of DNA polymerase beta that enhances alkylating agent and PARG inhibition induced cytotoxicity
- PMID: 38897003
- PMCID: PMC11239280
- DOI: 10.1016/j.dnarep.2024.103700
Oncometabolite 2-hydroxyglutarate suppresses basal protein levels of DNA polymerase beta that enhances alkylating agent and PARG inhibition induced cytotoxicity
Abstract
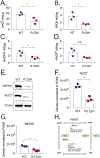
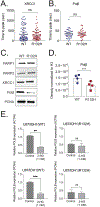
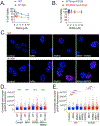
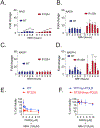
Mutations in isocitrate dehydrogenase isoform 1 (IDH1) are primarily found in secondary glioblastoma (GBM) and low-grade glioma but are rare in primary GBM. The standard treatment for GBM includes radiation combined with temozolomide, an alkylating agent. Fortunately, IDH1 mutant gliomas are sensitive to this treatment, resulting in a more favorable prognosis. However, it's estimated that up to 75 % of IDH1 mutant gliomas will progress to WHO grade IV over time and develop resistance to alkylating agents. Therefore, understanding the mechanism(s) by which IDH1 mutant gliomas confer sensitivity to alkylating agents is crucial for developing targeted chemotherapeutic approaches. The base excision repair (BER) pathway is responsible for repairing most base damage induced by alkylating agents. Defects in this pathway can lead to hypersensitivity to these agents due to unresolved DNA damage. The coordinated assembly and disassembly of BER protein complexes are essential for cell survival and for maintaining genomic integrity following alkylating agent exposure. These complexes rely on poly-ADP-ribose formation, an NAD+-dependent post-translational modification synthesized by PARP1 and PARP2 during the BER process. At the lesion site, poly-ADP-ribose facilitates the recruitment of XRCC1. This scaffold protein helps assemble BER proteins like DNA polymerase beta (Polβ), a bifunctional DNA polymerase containing both DNA synthesis and 5'-deoxyribose-phosphate lyase (5'dRP lyase) activity. Here, we confirm that IDH1 mutant glioma cells have defective NAD+ metabolism, but still produce sufficient nuclear NAD+ for robust PARP1 activation and BER complex formation in response to DNA damage. However, the overproduction of 2-hydroxyglutarate, an oncometabolite produced by the IDH1 R132H mutant protein, suppresses BER capacity by reducing Polβ protein levels. This defines a novel mechanism by which the IDH1 mutation in gliomas confers cellular sensitivity to alkylating agents and to inhibitors of the poly-ADP-ribose glycohydrolase, PARG.
Keywords: 2-hydroxyglutarate; Base excision repair; DNA polymerase beta; IDH1; NAD(+) metabolism; PARP1.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest R.W.S. is co-founder of Canal House Biosciences, LLC, is on the Scientific Advisory Board, and has an equity interest. Canal House Biosciences was not involved in the preparation of this study nor of this manuscript. The authors state that there is no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

