Microenvironment shapes small-cell lung cancer neuroendocrine states and presents therapeutic opportunities
- PMID: 38897168
- PMCID: PMC11228806
- DOI: 10.1016/j.xcrm.2024.101610
Microenvironment shapes small-cell lung cancer neuroendocrine states and presents therapeutic opportunities
Abstract
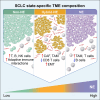
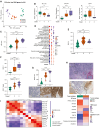
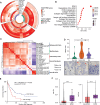
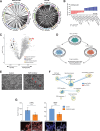
Small-cell lung cancer (SCLC) is the most fatal form of lung cancer. Intratumoral heterogeneity, marked by neuroendocrine (NE) and non-neuroendocrine (non-NE) cell states, defines SCLC, but the cell-extrinsic drivers of SCLC plasticity are poorly understood. To map the landscape of SCLC tumor microenvironment (TME), we apply spatially resolved transcriptomics and quantitative mass spectrometry-based proteomics to metastatic SCLC tumors obtained via rapid autopsy. The phenotype and overall composition of non-malignant cells in the TME exhibit substantial variability, closely mirroring the tumor phenotype, suggesting TME-driven reprogramming of NE cell states. We identify cancer-associated fibroblasts (CAFs) as a crucial element of SCLC TME heterogeneity, contributing to immune exclusion, and predicting exceptionally poor prognosis. Our work provides a comprehensive map of SCLC tumor and TME ecosystems, emphasizing their pivotal role in SCLC's adaptable nature, opening possibilities for reprogramming the TME-tumor communications that shape SCLC tumor states.
Keywords: cancer-associated fibroblasts; intercellular communication; rapid research autopsy; small-cell lung cancer; spatial transcriptomics; tumor heterogeneity; tumor microenvironment.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests A.T. received grants to NCI from EMD Serono Research & Development, AstraZeneca, Gilead Sciences, and ProLynx during the conduct of the study.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

