Therapeutic potential of co-signaling receptor modulation in hepatitis B
- PMID: 38897196
- PMCID: PMC11290321
- DOI: 10.1016/j.cell.2024.05.038
Therapeutic potential of co-signaling receptor modulation in hepatitis B
Abstract
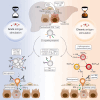
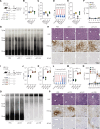
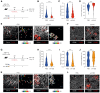
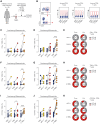
Reversing CD8+ T cell dysfunction is crucial in treating chronic hepatitis B virus (HBV) infection, yet specific molecular targets remain unclear. Our study analyzed co-signaling receptors during hepatocellular priming and traced the trajectory and fate of dysfunctional HBV-specific CD8+ T cells. Early on, these cells upregulate PD-1, CTLA-4, LAG-3, OX40, 4-1BB, and ICOS. While blocking co-inhibitory receptors had minimal effect, activating 4-1BB and OX40 converted them into antiviral effectors. Prolonged stimulation led to a self-renewing, long-lived, heterogeneous population with a unique transcriptional profile. This includes dysfunctional progenitor/stem-like (TSL) cells and two distinct dysfunctional tissue-resident memory (TRM) populations. While 4-1BB expression is ubiquitously maintained, OX40 expression is limited to TSL. In chronic settings, only 4-1BB stimulation conferred antiviral activity. In HBeAg+ chronic patients, 4-1BB activation showed the highest potential to rejuvenate dysfunctional CD8+ T cells. Targeting all dysfunctional T cells, rather than only stem-like precursors, holds promise for treating chronic HBV infection.
Keywords: 4-1BB; CD8(+) T cells; OX40; T cell dysfunction; chronic viral infection; hepatitis B virus; immunotherapy; liver.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M. Iannacone participates in advisory boards/consultantship for Asher Biotherapeutics, GentiBio, BlueJay Therapeutics, and Aligos Therapeutics. L.G.G. participates in boards/consultantship for Genenta Science, Epsilen Bio, Aligos Therapeutics, Medicxi, Chroma Medicine, and Ananda Immunotherapies.
Figures










References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

