Unravelling the mechanisms of antibiotic and heavy metal resistance co-selection in environmental bacteria
- PMID: 38897736
- PMCID: PMC11253441
- DOI: 10.1093/femsre/fuae017
Unravelling the mechanisms of antibiotic and heavy metal resistance co-selection in environmental bacteria
Abstract
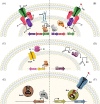
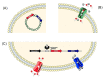
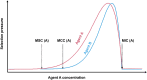
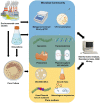
The co-selective pressure of heavy metals is a contributor to the dissemination and persistence of antibiotic resistance genes in environmental reservoirs. The overlapping range of antibiotic and metal contamination and similarities in their resistance mechanisms point to an intertwined evolutionary history. Metal resistance genes are known to be genetically linked to antibiotic resistance genes, with plasmids, transposons, and integrons involved in the assembly and horizontal transfer of the resistance elements. Models of co-selection between metals and antibiotics have been proposed, however, the molecular aspects of these phenomena are in many cases not defined or quantified and the importance of specific metals, environments, bacterial taxa, mobile genetic elements, and other abiotic or biotic conditions are not clear. Co-resistance is often suggested as a dominant mechanism, but interpretations are beset with correlational bias. Proof of principle examples of cross-resistance and co-regulation has been described but more in-depth characterizations are needed, using methodologies that confirm the functional expression of resistance genes and that connect genes with specific bacterial hosts. Here, we comprehensively evaluate the recent evidence for different models of co-selection from pure culture and metagenomic studies in environmental contexts and we highlight outstanding questions.
Keywords: antibiotics; evolution; metals; plasmids; resistance; selection.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures
References
-
- Ali H, Khan E. What are heavy metals? Long-standing controversy over the scientific use of the term ‘heavy metals’—proposal of a comprehensive definition. Toxicol Environ Chem. 2018;100:6–19.
-
- Allignet J, Loncle V, Simenel C et al. Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene. 1993;130:91–8. - PubMed
-
- Arsene-Ploetze F, Chiboub O, Lievremont D et al. Adaptation in toxic environments: comparative genomics of loci carrying antibiotic resistance genes derived from acid mine drainage waters. Environ Sci Pollut Res. 2018;25:1470–83. - PubMed
-
- Arya S, Williams A, Reina SV et al. Towards a general model for predicting minimal metal concentrations co-selecting for antibiotic resistance plasmids. Environ Pollut. 2021;275:116602. - PubMed
-
- Babakhani S, Oloomi M. Transposons: the agents of antibiotic resistance in bacteria. J Basic Microbiol. 2018;58:905–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

