Therapy-induced secretion of spliceosomal components mediates pro-survival crosstalk between ovarian cancer cells
- PMID: 38898005
- PMCID: PMC11187153
- DOI: 10.1038/s41467-024-49512-6
Therapy-induced secretion of spliceosomal components mediates pro-survival crosstalk between ovarian cancer cells
Erratum in
-
Author Correction: Therapy-induced secretion of spliceosomal components mediates pro-survival crosstalk between ovarian cancer cells.Nat Commun. 2024 Jul 31;15(1):6450. doi: 10.1038/s41467-024-50958-x. Nat Commun. 2024. PMID: 39085258 Free PMC article. No abstract available.
Abstract
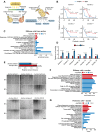
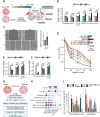
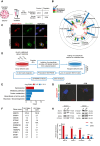
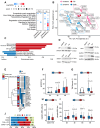
Ovarian cancer often develops resistance to conventional therapies, hampering their effectiveness. Here, using ex vivo paired ovarian cancer ascites obtained before and after chemotherapy and in vitro therapy-induced secretomes, we show that molecules secreted by ovarian cancer cells upon therapy promote cisplatin resistance and enhance DNA damage repair in recipient cancer cells. Even a short-term incubation of chemonaive ovarian cancer cells with therapy-induced secretomes induces changes resembling those that are observed in chemoresistant patient-derived tumor cells after long-term therapy. Using integrative omics techniques, we find that both ex vivo and in vitro therapy-induced secretomes are enriched with spliceosomal components, which relocalize from the nucleus to the cytoplasm and subsequently into the extracellular vesicles upon treatment. We demonstrate that these molecules substantially contribute to the phenotypic effects of therapy-induced secretomes. Thus, SNU13 and SYNCRIP spliceosomal proteins promote therapy resistance, while the exogenous U12 and U6atac snRNAs stimulate tumor growth. These findings demonstrate the significance of spliceosomal network perturbation during therapy and further highlight that extracellular signaling might be a key factor contributing to the emergence of ovarian cancer therapy resistance.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Xiao, L., Tang, J., Li, W., Xu, X. & Zhang, H. Improved prognosis for recurrent epithelial ovarian cancer by early diagnosis and 125I seeds implantation during suboptimal secondary cytoreductive surgery: a case report and literature review. J. Ovarian Res.13, 141 (2020). 10.1186/s13048-020-00744-2 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

