Mitochondrial division inhibitor (mdivi-1) induces extracellular matrix (ECM)-detachment of viable breast cancer cells by a DRP1-independent mechanism
- PMID: 38898058
- PMCID: PMC11187114
- DOI: 10.1038/s41598-024-64228-9
Mitochondrial division inhibitor (mdivi-1) induces extracellular matrix (ECM)-detachment of viable breast cancer cells by a DRP1-independent mechanism
Abstract
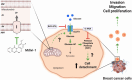
Increasing evidence supports the hypothesis that cancer progression is under mitochondrial control. Mitochondrial fission plays a pivotal role in the maintenance of cancer cell homeostasis. The inhibition of DRP1, the main regulator of mitochondrial fission, with the mitochondrial division inhibitor (mdivi-1) had been associated with cancer cell sensitivity to chemotherapeutics and decrease proliferation. Here, using breast cancer cells we find that mdivi-1 induces the detachment of the cells, leading to a bulk of floating cells that conserved their viability. Despite a decrease in their proliferative and clonogenic capabilities, these floating cells maintain the capacity to re-adhere upon re-seeding and retain their migratory and invasive potential. Interestingly, the cell detachment induced by mdivi-1 is independent of DRP1 but relies on inhibition of mitochondrial complex I. Furthermore, mdivi-1 induces cell detachment rely on glucose and the pentose phosphate pathway. Our data evidence a novel DRP1-independent effect of mdivi-1 in the attachment of cancer cells. The generation of floating viable cells restricts the use of mdivi-1 as a therapeutic agent and demonstrates that mdivi-1 effect on cancer cells are more complex than anticipated.
Keywords: Cancer; Cell detachment; Mdivi-1; Metabolism; Mitochondrial complex I.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Mitochondrial division inhibitor (mdivi-1) decreases oxidative metabolism in cancer.Br J Cancer. 2020 Apr;122(9):1288-1297. doi: 10.1038/s41416-020-0778-x. Epub 2020 Mar 9. Br J Cancer. 2020. PMID: 32147668 Free PMC article.
-
Mitochondrial division inhibitor 1 reduces dynamin-related protein 1 and mitochondrial fission activity.Hum Mol Genet. 2019 Jan 15;28(2):177-199. doi: 10.1093/hmg/ddy335. Hum Mol Genet. 2019. PMID: 30239719 Free PMC article.
-
Mitochondrial division inhibitor 1 disrupts oligodendrocyte Ca2+ homeostasis and mitochondrial function.Glia. 2020 Sep;68(9):1743-1756. doi: 10.1002/glia.23802. Epub 2020 Feb 14. Glia. 2020. PMID: 32060978
-
To mdivi-1 or not to mdivi-1: Is that the question?Dev Neurobiol. 2017 Nov;77(11):1260-1268. doi: 10.1002/dneu.22519. Epub 2017 Aug 30. Dev Neurobiol. 2017. PMID: 28842943 Free PMC article. Review.
-
Mdivi-1 and mitochondrial fission: recent insights from fungal pathogens.Curr Genet. 2019 Aug;65(4):837-845. doi: 10.1007/s00294-019-00942-6. Epub 2019 Feb 19. Curr Genet. 2019. PMID: 30783741 Free PMC article. Review.
Cited by
-
A review of the pathogenesis of mitochondria in breast cancer and progress of targeting mitochondria for breast cancer treatment.J Transl Med. 2025 Jan 15;23(1):70. doi: 10.1186/s12967-025-06077-2. J Transl Med. 2025. PMID: 39815317 Free PMC article. Review.
-
Mitophagy in the mechanisms of treatment resistance in solid tumors.Oncol Rev. 2025 Jul 21;19:1607983. doi: 10.3389/or.2025.1607983. eCollection 2025. Oncol Rev. 2025. PMID: 40761374 Free PMC article. Review.
-
Ginsenoside Rh1 Alleviates Allergic Rhinitis by Mediating Mitochondrial Autophagy via Activation of the AMPK/ULK1/FUNDC1 Pathway.Food Sci Nutr. 2025 Jun 17;13(6):e70464. doi: 10.1002/fsn3.70464. eCollection 2025 Jun. Food Sci Nutr. 2025. PMID: 40535916 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 3220604/Fondo Nacional de Desarrollo Científico y Tecnológico
- 21232288/Fondo Nacional de Desarrollo Científico y Tecnológico
- 21212019/Fondo Nacional de Desarrollo Científico y Tecnológico
- 3220593/Fondo Nacional de Desarrollo Científico y Tecnológico
- 3230273/Fondo Nacional de Desarrollo Científico y Tecnológico
- 1200255/Fondo Nacional de Desarrollo Científico y Tecnológico
- 21181428/Fondo Nacional de Desarrollo Científico y Tecnológico
- 1230983/Fondo Nacional de Desarrollo Científico y Tecnológico
- 11221017/Fondo Nacional de Desarrollo Científico y Tecnológico
- 15150012/Fondo de Financiamiento de Centros de Investigación en Áreas Prioritarias
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

