High capacity clinical SARS-CoV-2 molecular testing using combinatorial pooling
- PMID: 38898090
- PMCID: PMC11187214
- DOI: 10.1038/s43856-024-00531-w
High capacity clinical SARS-CoV-2 molecular testing using combinatorial pooling
Abstract
Background: The SARS-CoV-2 pandemic led to unprecedented testing demands, causing major testing delays globally. One strategy used for increasing testing capacity was pooled-testing, using a two-stage technique first introduced during WWII. However, such traditional pooled testing was used in practice only when positivity rates were below 2%.
Methods: Here we report the development, validation and clinical application of P-BEST - a single-stage pooled-testing strategy that was approved for clinical use in Israel.
Results: P-BEST is clinically validated using 3636 side-by-side tests and is able to correctly detect all positive samples and accurately estimate their Ct value. Following regulatory approval by the Israeli Ministry of Health, P-BEST was used in 2021 to clinically test 837,138 samples using 270,095 PCR tests - a 3.1fold reduction in the number of tests. This period includes the Alpha and Delta waves, when positivity rates exceeded 10%, rendering traditional pooling non-practical. We also describe a tablet-based solution that allows performing manual single-stage pooling in settings where liquid dispensing robots are not available.
Conclusions: Our data provides a proof-of-concept for large-scale clinical implementation of single-stage pooled-testing for continuous surveillance of multiple pathogens with reduced test costs, and as an important tool for increasing testing efficiency during pandemic outbreaks.
Plain language summary
Testing samples for SARS-CoV-2 is usually done on one sample at a time. However, the unprecedented demand for testing during the COVID-19 pandemic led to the adoption of pooled testing strategies, where samples are combined before being tested. This strategy requires two rounds: first, each pool of samples is tested, and then a second testing round is performed on individual samples from positive pools. We developed and implemented a pooling method for SARS-CoV-2 that requires a single round of testing, thus enabling the shorter turnaround times required during a pandemic. The method was approved for clinical use in Israel and was used to successfully test 837,138 clinical samples using fewer than a third of the tests usually required. Our study provides a blueprint for rapid implementation of efficient high-throughput testing in future pandemics.
© 2024. The Author(s).
Conflict of interest statement
AP, NS and TH are co-founders and shareholders in Poold Diagnostics.
Figures

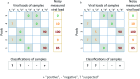

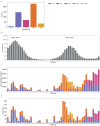

References
-
- Ritchie, H. et al. Coronavirus Pandemic (COVID-19). Our World in Data (2020).
-
- FDA U.S Food and Drug Administration, F. D. A. Molecular-Based Diagnostic Tests Authorized for Emergency Use for Coronavirus Disease 2019 (COVID-19), (2021). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

