Multiscale topology classifies cells in subcellular spatial transcriptomics
- PMID: 38898271
- PMCID: PMC11208150
- DOI: 10.1038/s41586-024-07563-1
Multiscale topology classifies cells in subcellular spatial transcriptomics
Abstract
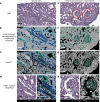
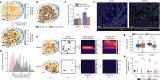
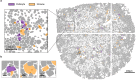
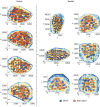
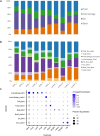
Spatial transcriptomics measures in situ gene expression at millions of locations within a tissue1, hitherto with some trade-off between transcriptome depth, spatial resolution and sample size2. Although integration of image-based segmentation has enabled impactful work in this context, it is limited by imaging quality and tissue heterogeneity. By contrast, recent array-based technologies offer the ability to measure the entire transcriptome at subcellular resolution across large samples3-6. Presently, there exist no approaches for cell type identification that directly leverage this information to annotate individual cells. Here we propose a multiscale approach to automatically classify cell types at this subcellular level, using both transcriptomic information and spatial context. We showcase this on both targeted and whole-transcriptome spatial platforms, improving cell classification and morphology for human kidney tissue and pinpointing individual sparsely distributed renal mouse immune cells without reliance on image data. By integrating these predictions into a topological pipeline based on multiparameter persistent homology7-9, we identify cell spatial relationships characteristic of a mouse model of lupus nephritis, which we validate experimentally by immunofluorescence. The proposed framework readily generalizes to new platforms, providing a comprehensive pipeline bridging different levels of biological organization from genes through to tissues.
© 2024. The Author(s).
Conflict of interest statement
The chip, procedure and applications of Stereo-seq are covered in a patent application (application no. PCT/CN2020/090340; under prosecution at national stage; applicants are BGI Shenzhen, MGI Tech; inventors are A. Chen, X. Xun, J. Yang, L. Liu, O. Wang, Y. Li, S. Liao, G. Tang, Y. Jiang, C. Xu, M. Ni, W. Zhang, R. Drmanac and S. Drmanac). Z.S., Y.X., Y.A., N.Z. and Y.H. are employees of BGI and have stock holdings in BGI. The remaining authors declare no competing interests.
Figures





References
-
- Nagendran M, et al. Visium HD enables spatially resolved, single-cell scale resolution mapping of FFPE human breast cancer tissue. J. Immunother. Cancer. 2023;11:A1620.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

