dsRNA formation leads to preferential nuclear export and gene expression
- PMID: 38898279
- PMCID: PMC11236707
- DOI: 10.1038/s41586-024-07576-w
dsRNA formation leads to preferential nuclear export and gene expression
Abstract
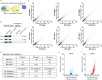
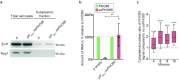
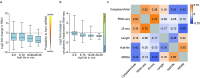
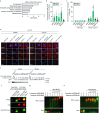
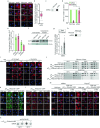
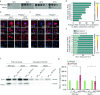
When mRNAs have been transcribed and processed in the nucleus, they are exported to the cytoplasm for translation. This export is mediated by the export receptor heterodimer Mex67-Mtr2 in the yeast Saccharomyces cerevisiae (TAP-p15 in humans)1,2. Interestingly, many long non-coding RNAs (lncRNAs) also leave the nucleus but it is currently unclear why they move to the cytoplasm3. Here we show that antisense RNAs (asRNAs) accelerate mRNA export by annealing with their sense counterparts through the helicase Dbp2. These double-stranded RNAs (dsRNAs) dominate export compared with single-stranded RNAs (ssRNAs) because they have a higher capacity and affinity for the export receptor Mex67. In this way, asRNAs boost gene expression, which is beneficial for cells. This is particularly important when the expression program changes. Consequently, the degradation of dsRNA, or the prevention of its formation, is toxic for cells. This mechanism illuminates the general cellular occurrence of asRNAs and explains their nuclear export.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

