Characterization and Genome Analysis of the Delftia lacustris Strain LzhVag01 Isolated from Vaginal Discharge
- PMID: 38898312
- PMCID: PMC11186869
- DOI: 10.1007/s00284-024-03758-x
Characterization and Genome Analysis of the Delftia lacustris Strain LzhVag01 Isolated from Vaginal Discharge
Abstract
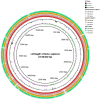
Delftia has been separated from freshwater, sludge, and soil and has emerged as a novel opportunistic pathogen in the female vagina. However, the genomic characteristics, pathogenicity, and biotechnological properties still need to be comprehensively investigated. In this study, a Delftia strain was isolated from the vaginal discharge of a 43-year-old female with histologically confirmed cervical intraepithelial neoplasm (CIN III), followed by whole-genome sequencing. Phylogenetic analysis and average nucleotide identity (ANI) analysis demonstrated that it belongs to Delftia lacustris, named D. lacustris strain LzhVag01. LzhVag01 was sensitive to β-lactams, macrolides, and tetracyclines but exhibited resistance to lincoamines, nitroimidazoles, aminoglycosides, and fluoroquinolones. Its genome is a single, circular chromosome of 6,740,460 bp with an average GC content of 66.59%. Whole-genome analysis identified 16 antibiotic resistance-related genes, which match the antimicrobial susceptibility profile of this strain, and 11 potential virulence genes. These pathogenic factors may contribute to its colonization in the vaginal environment and its adaptation and accelerate the progression of cervical cancer. This study sequenced and characterized the whole-genome of Delftia lacustris isolated from vaginal discharge, which provides investigators and clinicians with valuable insights into this uncommon species.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Genomic Analysis of Delftia tsuruhatensis Strain TR1180 Isolated From A Patient From China With In4-Like Integron-Associated Antimicrobial Resistance.Front Cell Infect Microbiol. 2021 Jun 17;11:663933. doi: 10.3389/fcimb.2021.663933. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34222039 Free PMC article.
-
Characterization and Genomic Analysis of Quinolone-Resistant Delftia sp. 670 Isolated from a Patient Who Died from Severe Pneumonia.Curr Microbiol. 2015 Jul;71(1):54-61. doi: 10.1007/s00284-015-0818-6. Epub 2015 May 3. Curr Microbiol. 2015. PMID: 25935202
-
Pan-Genome Analysis of Delftia tsuruhatensis Reveals Important Traits Concerning the Genetic Diversity, Pathogenicity, and Biotechnological Properties of the Species.Microbiol Spectr. 2022 Apr 27;10(2):e0207221. doi: 10.1128/spectrum.02072-21. Epub 2022 Mar 1. Microbiol Spectr. 2022. PMID: 35230132 Free PMC article.
-
Genome sequencing reveals mechanisms for heavy metal resistance and polycyclic aromatic hydrocarbon degradation in Delftia lacustris strain LZ-C.Ecotoxicology. 2016 Jan;25(1):234-47. doi: 10.1007/s10646-015-1583-9. Epub 2015 Nov 20. Ecotoxicology. 2016. PMID: 26589947
-
Delftia lacustris septicemia in a pheochromocytoma patient: case report and literature review.Infect Dis (Lond). 2015 May;47(5):349-53. doi: 10.3109/00365548.2014.993422. Epub 2015 Feb 24. Infect Dis (Lond). 2015. PMID: 25712727 Review.
Cited by
-
Alterations of the paired maternal vaginal microbiome and neonatal meconium microbiome in vulvovaginal candidiasis positive pregnant women.Front Cell Infect Microbiol. 2024 Dec 13;14:1480200. doi: 10.3389/fcimb.2024.1480200. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39735259 Free PMC article.
-
Assessment of the wheat growth-promoting potential of Delftia lacustris strain NSC through genomic and physiological characterization.Front Microbiol. 2025 Jun 16;16:1576536. doi: 10.3389/fmicb.2025.1576536. eCollection 2025. Front Microbiol. 2025. PMID: 40589583 Free PMC article.
-
Laundry Isolate Delftia sp. UBM14 Capable of Biodegrading Industrially Relevant Aminophosphonates.Microorganisms. 2024 Aug 13;12(8):1664. doi: 10.3390/microorganisms12081664. Microorganisms. 2024. PMID: 39203506 Free PMC article.
References
-
- Yin Z, Liu X, Qian C et al (2022) Pan-genome analysis of Delftia tsuruhatensis reveals important traits concerning the genetic diversity, pathogenicity, and biotechnological properties of the species. Microbiol Spectr 10:e02072-e2121. 10.1128/spectrum.02072-21 10.1128/spectrum.02072-21 - DOI - PMC - PubMed
-
- Jorgensen NOG, Brandt KK, Nybroe O, Hansen M (2009) Delftia lacustris sp. nov., a peptidoglycan-degrading bacterium from fresh water, and emended description of Delftia tsuruhatensis as a peptidoglycan-degrading bacterium. Int J Syst Evol Microbiol 59:2195–2199. 10.1099/ijs.0.008375-0 10.1099/ijs.0.008375-0 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

