Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome
- PMID: 38898526
- PMCID: PMC11186191
- DOI: 10.1186/s12977-024-00645-y
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome
Abstract
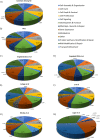
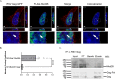
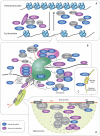
Retroviruses exploit host proteins to assemble and release virions from infected cells. Previously, most studies focused on interacting partners of retroviral Gag proteins that localize to the cytoplasm or plasma membrane. Given that several full-length Gag proteins have been found in the nucleus, identifying the Gag-nuclear interactome has high potential for novel findings involving previously unknown host processes. Here we systematically compared nuclear factors identified in published HIV-1 proteomic studies and performed our own mass spectrometry analysis using affinity-tagged HIV-1 and RSV Gag proteins mixed with nuclear extracts. We identified 57 nuclear proteins in common between HIV-1 and RSV Gag, and a set of nuclear proteins present in our analysis and ≥ 1 of the published HIV-1 datasets. Many proteins were associated with nuclear processes which could have functional consequences for viral replication, including transcription initiation/elongation/termination, RNA processing, splicing, and chromatin remodeling. Examples include facilitating chromatin remodeling to expose the integrated provirus, promoting expression of viral genes, repressing the transcription of antagonistic cellular genes, preventing splicing of viral RNA, altering splicing of cellular RNAs, or influencing viral or host RNA folding or RNA nuclear export. Many proteins in our pulldowns common to RSV and HIV-1 Gag are critical for transcription, including PolR2B, the second largest subunit of RNA polymerase II (RNAPII), and LEO1, a PAF1C complex member that regulates transcriptional elongation, supporting the possibility that Gag influences the host transcription profile to aid the virus. Through the interaction of RSV and HIV-1 Gag with splicing-related proteins CBLL1, HNRNPH3, TRA2B, PTBP1 and U2AF1, we speculate that Gag could enhance unspliced viral RNA production for translation and packaging. To validate one putative hit, we demonstrated an interaction of RSV Gag with Mediator complex member Med26, required for RNA polymerase II-mediated transcription. Although 57 host proteins interacted with both Gag proteins, unique host proteins belonging to each interactome dataset were identified. These results provide a strong premise for future functional studies to investigate roles for these nuclear host factors that may have shared functions in the biology of both retroviruses, as well as functions specific to RSV and HIV-1, given their distinctive hosts and molecular pathology.
Keywords: HIV-1; Mass spectrometry; Proteomics; Retroviruses; Rous sarcoma virus.
© 2024. The Author(s).
Conflict of interest statement
Leslie Parent is an Associate Editor for Retrovirology. The authors declare no conflict of interest.
Figures


Update of
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.bioRxiv [Preprint]. 2024 Mar 6:2024.01.18.575255. doi: 10.1101/2024.01.18.575255. bioRxiv. 2024. Update in: Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y. PMID: 38293010 Free PMC article. Updated. Preprint.
Similar articles
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.bioRxiv [Preprint]. 2024 Mar 6:2024.01.18.575255. doi: 10.1101/2024.01.18.575255. bioRxiv. 2024. Update in: Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y. PMID: 38293010 Free PMC article. Updated. Preprint.
-
Visualizing Association of the Retroviral Gag Protein with Unspliced Viral RNA in the Nucleus.mBio. 2020 Apr 7;11(2):e00524-20. doi: 10.1128/mBio.00524-20. mBio. 2020. PMID: 32265329 Free PMC article.
-
An Infectious Rous Sarcoma Virus Gag Mutant That Is Defective in Nuclear Cycling.J Virol. 2021 Sep 27;95(20):e0064821. doi: 10.1128/JVI.00648-21. Epub 2021 Jul 28. J Virol. 2021. PMID: 34319154 Free PMC article.
-
Nuclear trafficking of retroviral RNAs and Gag proteins during late steps of replication.Viruses. 2013 Nov 18;5(11):2767-95. doi: 10.3390/v5112767. Viruses. 2013. PMID: 24253283 Free PMC article. Review.
-
The structure and function of the rous sarcoma virus RNA stability element.J Cell Biochem. 2011 Nov;112(11):3085-92. doi: 10.1002/jcb.23272. J Cell Biochem. 2011. PMID: 21769913 Free PMC article. Review.
Cited by
-
Dynamic interactions of retroviral Gag condensates with nascent viral RNA at transcriptional burst sites: implications for genomic RNA packaging.bioRxiv [Preprint]. 2025 Jun 2:2025.01.11.632546. doi: 10.1101/2025.01.11.632546. bioRxiv. 2025. PMID: 39829876 Free PMC article. Preprint.
-
Two peas in a pod: retroviral RNA dimers organize Gag-RNA nanoclusters with novel biophysical properties.bioRxiv [Preprint]. 2025 Jun 11:2025.06.09.658653. doi: 10.1101/2025.06.09.658653. bioRxiv. 2025. Update in: Int J Mol Sci. 2025 Jun 13;26(12):5679. doi: 10.3390/ijms26125679. PMID: 40661432 Free PMC article. Updated. Preprint.
-
Role of the Psi Packaging Signal and Dimerization Initiation Sequence in the Organization of Rous Sarcoma Virus Gag-gRNA Co-Condensates.Viruses. 2025 Jan 13;17(1):97. doi: 10.3390/v17010097. Viruses. 2025. PMID: 39861886 Free PMC article.
-
Two Peas in a Pod: Retroviral RNA Dimers Organize Gag-RNA Nanoclusters with Novel Biophysical Properties.Int J Mol Sci. 2025 Jun 13;26(12):5679. doi: 10.3390/ijms26125679. Int J Mol Sci. 2025. PMID: 40565141 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

