Deep Learning-Based Glucose Prediction Models: A Guide for Practitioners and a Curated Dataset for Improved Diabetes Management
- PMID: 38899015
- PMCID: PMC11186642
- DOI: 10.1109/OJEMB.2024.3365290
Deep Learning-Based Glucose Prediction Models: A Guide for Practitioners and a Curated Dataset for Improved Diabetes Management
Abstract
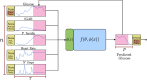
Accurate short- and mid-term blood glucose predictions are crucial for patients with diabetes struggling to maintain healthy glucose levels, as well as for individuals at risk of developing the disease. Consequently, numerous efforts from the scientific community have focused on developing predictive models for glucose levels. This study harnesses physiological data collected from wearable sensors to construct a series of data-driven models based on deep learning approaches. We systematically compare these models to offer insights for practitioners and researchers venturing into glucose prediction using deep learning techniques. Key questions addressed in this work encompass the comparison of various deep learning architectures for this task, determining the optimal set of input variables for accurate glucose prediction, comparing population-wide, fine-tuned, and personalized models, and assessing the impact of an individual's data volume on model performance. Additionally, as part of our outcomes, we introduce a meticulously curated dataset inclusive of data from both healthy individuals and those with diabetes, recorded in free-living conditions. This dataset aims to foster research in this domain and facilitate equitable comparisons among researchers.
Keywords: Diabetes; Glucose prediction; deep learning; transfer learning.
© 2024 The Authors.
Figures




References
-
- Roglic G. et al. , “WHO global report on diabetes: A summary,” Int. J. Noncommunicable Dis., vol. 1, no. 1, pp. 3–8, 2016.
-
- Saeedi P. et al. , “Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the international diabetes federation diabetes atlas,” Diabetes Res. Clin. Pract., vol. 157, 2019, Art. no. 107843. - PubMed
-
- Gorst C. et al. , “Long-term glycemic variability and risk of adverse outcomes: A systematic review and meta-analysis,” Diabetes Care, vol. 38, no. 12, pp. 2354–2369, 2015. - PubMed
-
- Huyett L. M., Dassau E., Zisser H. C., and Doyle F. J., “Glucose sensor dynamics and the artificial pancreas: The impact of lag on sensor measurement and controller performance,” IEEE Control Syst. Mag., vol. 38, no. 1, pp. 30–46, Feb. 2018.

