Interferon and B-cell Signatures Inform Precision Medicine in Lupus Nephritis
- PMID: 38899167
- PMCID: PMC11184261
- DOI: 10.1016/j.ekir.2024.03.014
Interferon and B-cell Signatures Inform Precision Medicine in Lupus Nephritis
Abstract
Introduction: Current therapeutic management of lupus nephritis (LN) fails to induce long-term remission in over 50% of patients, highlighting the urgent need for additional options.
Methods: We analyzed differentially expressed genes (DEGs) in peripheral blood from patients with active LN (n = 41) and active nonrenal lupus (n = 62) versus healthy controls (HCs) (n = 497) from the European PRECISESADS project (NTC02890121), and dysregulated gene modules in a discovery (n = 26) and a replication (n = 15) set of active LN cases.
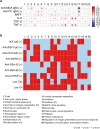
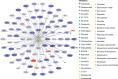
Results: Replicated gene modules qualified for correlation analyses with serologic markers, and regulatory network and druggability analysis. Unsupervised coexpression network analysis revealed 20 dysregulated gene modules and stratified the active LN population into 3 distinct subgroups. These subgroups were characterized by low, intermediate, and high interferon (IFN) signatures, with differential dysregulation of the "B cell" and "plasma cells/Ig" modules. Drugs annotated to the IFN network included CC-motif chemokine receptor 1 (CCR1) inhibitors, programmed death-ligand 1 (PD-L1) inhibitors, and irinotecan; whereas the anti-CD38 daratumumab and proteasome inhibitor bortezomib showed potential for counteracting the "plasma cells/Ig" signature. In silico analysis demonstrated the low-IFN subgroup to benefit from calcineurin inhibition and the intermediate-IFN subgroup from B-cell targeted therapies. High-IFN patients exhibited greater anticipated response to anifrolumab whereas daratumumab appeared beneficial to the intermediate-IFN and high-IFN subgroups.
Conclusion: IFN upregulation and B and plasma cell gene dysregulation patterns revealed 3 subgroups of LN, which may not necessarily represent distinct disease phenotypes but rather phases of the inflammatory processes during a renal flare, providing a conceptual framework for precision medicine in LN.
Keywords: biologics; druggability; lupus nephritis; precision medicine; systemic lupus erythematosus; trancriptomics; transcriptome.
© 2024 International Society of Nephrology. Published by Elsevier Inc.
Figures




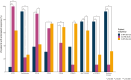

References
-
- Fanouriakis A., Kostopoulou M., Cheema K., et al. 2019 Update of the Joint European League Against Rheumatism and European Renal Association-European Dialysis and Transplant Association (EULAR/ERA-EDTA) recommendations for the management of lupus nephritis. Ann Rheum Dis. 2020;79:713–723. doi: 10.1136/annrheumdis-2020-216924. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

