An initial HOPS-mediated fusion event is critical for autophagosome transport initiation from the axon terminal
- PMID: 38899385
- PMCID: PMC11423661
- DOI: 10.1080/15548627.2024.2366122
An initial HOPS-mediated fusion event is critical for autophagosome transport initiation from the axon terminal
Abstract
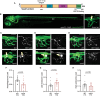
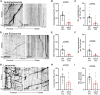
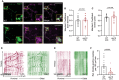
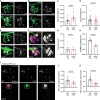
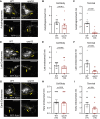
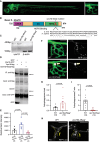
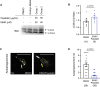
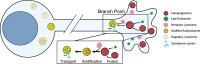
In neurons, macroautophagy/autophagy is a frequent and critical process. In the axon, autophagy begins in the axon terminal, where most nascent autophagosomes form. After formation, autophagosomes must initiate transport to exit the axon terminal and move toward the cell body via retrograde transport. During retrograde transport these autophagosomes mature through repetitive fusion events. Complete lysosomal cargo degradation occurs largely in the cell body. The precipitating events to stimulate retrograde autophagosome transport have been debated but their importance is clear: disrupting neuronal autophagy or autophagosome transport is detrimental to neuronal health and function. We have identified the HOPS complex as essential for early autophagosome maturation and consequent initiation of retrograde transport from the axon terminal. In yeast and mammalian cells, HOPS controls fusion between autophagosomes and late endosomes with lysosomes. Using zebrafish strains with loss-of-function mutations in vps18 and vps41, core components of the HOPS complex, we found that disruption of HOPS eliminates autophagosome maturation and disrupts retrograde autophagosome transport initiation from the axon terminal. We confirmed this phenotype was due to loss of HOPS complex formation using an endogenous deletion of the HOPS binding domain in Vps18. Finally, using pharmacological inhibition of lysosomal proteases, we show that initiation of autophagosome retrograde transport requires autophagosome maturation. Together, our data demonstrate that HOPS-mediated fusion events are critical for retrograde autophagosome transport initiation through promoting autophagosome maturation. This reveals critical roles for the HOPS complex in neuronal autophagy which deepens our understanding of the cellular pathology of HOPS-complex linked neurodegenerative diseases.Abbreviations: CORVET: Class C core vacuole/endosome tethering; gRNA: guide RNA; HOPS: homotypic fusion and protein sorting; pLL: posterior lateral line; Vps18: VPS18 core subunit of CORVET and HOPS complexes; Vps41: VPS41 subunit of HOPS complex.
Keywords: Axon terminal; Vps18; autophagy; axonal transport; lysosome; neuron.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
Comprehensive knockout analysis of the RAB family small GTPases reveals an overlapping role of RAB2 and RAB14 in autophagosome maturation.Autophagy. 2025 Jan;21(1):21-36. doi: 10.1080/15548627.2024.2374699. Epub 2024 Jul 10. Autophagy. 2025. PMID: 38953305 Free PMC article.
-
Apache is a neuronal player in autophagy required for retrograde axonal transport of autophagosomes.Cell Mol Life Sci. 2024 Oct 5;81(1):416. doi: 10.1007/s00018-024-05441-7. Cell Mol Life Sci. 2024. PMID: 39367928 Free PMC article.
-
TBC1D4 antagonizes RAB2A-mediated autophagic and endocytic pathways.Autophagy. 2024 Nov;20(11):2426-2443. doi: 10.1080/15548627.2024.2367907. Epub 2024 Jul 4. Autophagy. 2024. PMID: 38964379 Free PMC article.
-
Molecular Mechanism of Autophagosome-Lysosome Fusion in Mammalian Cells.Cells. 2024 Mar 13;13(6):500. doi: 10.3390/cells13060500. Cells. 2024. PMID: 38534345 Free PMC article. Review.
-
Spatio-temporal processes in autophagosome-lysosome fusion.Med Rev (2021). 2025 Jun 19;5(4):297-317. doi: 10.1515/mr-2024-0095. eCollection 2025 Aug. Med Rev (2021). 2025. PMID: 40838105 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
