Structural investigations and antibacterial, antifungal and anticancer studies on zinc salicylaldimine complexes
- PMID: 38899770
- PMCID: PMC11370977
- DOI: 10.1080/17568919.2024.2363672
Structural investigations and antibacterial, antifungal and anticancer studies on zinc salicylaldimine complexes
Abstract
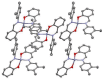
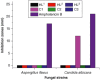
Aim: Zinc salicylaldimines may act as multidrug agents.Results: Three zinc salicylaldimines C1-C3 and respective ligands HL1-HL3 were examined for antimicrobial/anticancer drug action and C3 was structurally analyzed (tetrahedral, triclinic). Against two fungi, C1 inhibited Candida albicans with 12 mm (21 mm for amphotericin B). Among four bacteria, two ligands inhibited Staphylococcus aureus and Escherichia coli (9-10 mm), but the complexes inhibited all bacteria with 10-14 mm (21-26 mm for ampicillin). The half-maximal inhibitory concentrations for the ligands, complexes and doxorubicin were 195.5-310.7, 22.18-70.05 and 9.66 μM against cancerous MCF-7 cells and 186.4-199.9, 14.95-18.87 and 36.42 μM against normal BHK cells.Conclusion: The complexation produced pronounced enhancement in the ligand antimicrobial/anticancer activities, despite these activities are moderate comparing with standards.
Keywords: N and O chelation; anionic ligands; breast; diamagnetism; doxorubicin; kidney.
Plain language summary
[Box: see text].
Conflict of interest statement
The authors have no competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Figures




Similar articles
-
Discovery of new molecular hybrid derivatives with coumarin scaffold bearing pyrazole/oxadiazole moieties: Molecular docking, POM analyses, in silico pharmacokinetics and in vitro antimicrobial evaluation with identification of potent antitumor pharmacophore sites.Bioorg Chem. 2024 Dec;153:107761. doi: 10.1016/j.bioorg.2024.107761. Epub 2024 Aug 30. Bioorg Chem. 2024. PMID: 39241586
-
New bio-sensitive and biologically active single crystal of pyrimidine scaffold ligand and its gold and platinum complexes: DFT, antimicrobial, antioxidant, DNA interaction, molecular docking with DNA/BSA and anticancer studies.Bioorg Chem. 2018 Dec;81:144-156. doi: 10.1016/j.bioorg.2018.08.006. Epub 2018 Aug 11. Bioorg Chem. 2018. PMID: 30121002
-
Antimicrobial and Antioxidant Electrospun Membrane Incorporating Silver and Copper Complexes of Phenylboronic Acid-Functionalized 4,5-Diazafluorene Ligand.Arch Pharm (Weinheim). 2025 Aug;358(8):e70072. doi: 10.1002/ardp.70072. Arch Pharm (Weinheim). 2025. PMID: 40778687
-
A comprehensive review on anti-inflammatory, antibacterial, anticancer and antifungal properties of several bivalent transition metal complexes.Bioorg Chem. 2025 Jun 15;160:108422. doi: 10.1016/j.bioorg.2025.108422. Epub 2025 Apr 1. Bioorg Chem. 2025. PMID: 40187028 Review.
-
A review on coordination and biological properties of selenosemicarbazone.J Inorg Biochem. 2025 Nov;272:113020. doi: 10.1016/j.jinorgbio.2025.113020. Epub 2025 Aug 7. J Inorg Biochem. 2025. PMID: 40780005 Review.
References
-
- Krishnamoorthy P, Sathyadevi P, Muthiah PT, et al. Nickel and cobalt complexes of benzoic acid (2-hydroxy-benzylidene)-hydrazide ligand: synthesis, structure and comparative in vitro evaluations of biological perspectives. RSC Adv. 2012;2:12190–12203. doi: 10.1039/c2ra20597a - DOI
-
- Sano T, Nishio Y, Hamada Y, et al. Design of conjugated molecular materials for optoelectronics. J Mater Chem. 2000;10:157–161. doi: 10.1039/a903239h - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
