A comparative study of large language model-based zero-shot inference and task-specific supervised classification of breast cancer pathology reports
- PMID: 38900207
- PMCID: PMC11413420
- DOI: 10.1093/jamia/ocae146
A comparative study of large language model-based zero-shot inference and task-specific supervised classification of breast cancer pathology reports
Abstract
Objective: Although supervised machine learning is popular for information extraction from clinical notes, creating large annotated datasets requires extensive domain expertise and is time-consuming. Meanwhile, large language models (LLMs) have demonstrated promising transfer learning capability. In this study, we explored whether recent LLMs could reduce the need for large-scale data annotations.
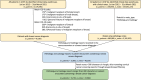
Materials and methods: We curated a dataset of 769 breast cancer pathology reports, manually labeled with 12 categories, to compare zero-shot classification capability of the following LLMs: GPT-4, GPT-3.5, Starling, and ClinicalCamel, with task-specific supervised classification performance of 3 models: random forests, long short-term memory networks with attention (LSTM-Att), and the UCSF-BERT model.
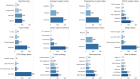
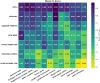
Results: Across all 12 tasks, the GPT-4 model performed either significantly better than or as well as the best supervised model, LSTM-Att (average macro F1-score of 0.86 vs 0.75), with advantage on tasks with high label imbalance. Other LLMs demonstrated poor performance. Frequent GPT-4 error categories included incorrect inferences from multiple samples and from history, and complex task design, and several LSTM-Att errors were related to poor generalization to the test set.
Discussion: On tasks where large annotated datasets cannot be easily collected, LLMs can reduce the burden of data labeling. However, if the use of LLMs is prohibitive, the use of simpler models with large annotated datasets can provide comparable results.
Conclusions: GPT-4 demonstrated the potential to speed up the execution of clinical NLP studies by reducing the need for large annotated datasets. This may increase the utilization of NLP-based variables and outcomes in clinical studies.
Keywords: breast cancer; electronic health records; large language models; natural language processing; pathology.
© The Author(s) 2024. Published by Oxford University Press on behalf of the American Medical Informatics Association.
Conflict of interest statement
M.S. reports no financial associations or conflicts of interest. T.Z. is a medical advisor and minor shareholder at OpenEvidence.com. D.M. is a consultant to Third Rock Ventures. Z.Z. reports no financial associations or conflicts of interest. A.W. is currently an employee of Abbott. Y.-N.Y. is currently an employee of City of Hope. Y.Q. is currently an employee of X-camp Academy. D.L. is currently an employee and minor shareholder of Johnson and Johnson and a co-founder and major shareholder of Synthez AI Corp. A.J.B. is a co-founder and consultant to Personalis and NuMedii; consultant to Mango Tree Corporation, and in the recent past, Samsung, 10x Genomics, Helix, Pathway Genomics, and Verinata (Illumina); has served on paid advisory panels or boards for Geisinger Health, Regenstrief Institute, Gerson Lehman Group, AlphaSights, Covance, Novartis, Genentech, and Merck, and Roche; is a shareholder in Personalis and NuMedii; is a minor shareholder in Apple, Meta (Facebook), Alphabet (Google), Microsoft, Amazon, Snap, 10x Genomics, Illumina, Regeneron, Sanofi, Pfizer, Royalty Pharma, Moderna, Sutro, Doximity, BioNtech, Invitae, Pacific Biosciences, Editas Medicine, Nuna Health, Assay Depot, and Vet24seven, and several other non-health related companies and mutual funds; and has received honoraria and travel reimbursement for invited talks from Johnson and Johnson, Roche, Genentech, Pfizer, Merck, Lilly, Takeda, Varian, Mars, Siemens, Optum, Abbott, Celgene, AstraZeneca, AbbVie, Westat, and many academic institutions, medical or disease specific foundations and associations, and health systems. A.J.B. receives royalty payments through Stanford University, for several patents and other disclosures licensed to NuMedii and Personalis. A.J.B.’s research has been funded by NIH, Peraton (as the prime on an NIH contract), Genentech, Johnson and Johnson, FDA, Robert Wood Johnson Foundation, Leon Lowenstein Foundation, Intervalien Foundation, Priscilla Chan and Mark Zuckerberg, the Barbara and Gerson Bakar Foundation, and in the recent past, the March of Dimes, Juvenile Diabetes Research Foundation, California Governor’s Office of Planning and Research, California Institute for Regenerative Medicine, L’Oreal, and Progenity. None of these entities had any bearing on this research or interpretation of the findings.
Figures

Update of
-
A comparative study of zero-shot inference with large language models and supervised modeling in breast cancer pathology classification.Res Sq [Preprint]. 2024 Feb 6:rs.3.rs-3914899. doi: 10.21203/rs.3.rs-3914899/v1. Res Sq. 2024. Update in: J Am Med Inform Assoc. 2024 Oct 1;31(10):2315-2327. doi: 10.1093/jamia/ocae146. PMID: 38405831 Free PMC article. Updated. Preprint.
References
-
- Brown T, Mann B, Ryder N, et al. Language models are few-shot learners. In: Larochelle H, Ranzato M, Hadsell R, et al., eds. Advances in Neural Information Processing Systems. Vol. 33. Curran Associates, Inc.; 2020:1877-1901.
-
- Kojima T, Gu S, Shane R, Matsuo M, Iwasawa Y. Y. . Large language models are zero-shot reasoners. In: Koyejo S, Mohamed S, Agarwal A, et al., eds. Adv Neural Inform Process Syst. 2022;35:22199-22213.
-
- Agrawal M, Hegselmann S, Lang H, Kim Y, Sontag D. Large language models are few-shot clinical information extractors. In: Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing 1998–2022 (Association for Computational Linguistics, Abu Dhabi, United Arab Emirates, 2022).

