Transcriptional regulation of CYR61 and CTGF by LM98: a synthetic YAP-TEAD inhibitor that targets in-vitro vasculogenic mimicry in glioblastoma cells
- PMID: 38900643
- PMCID: PMC11305628
- DOI: 10.1097/CAD.0000000000001627
Transcriptional regulation of CYR61 and CTGF by LM98: a synthetic YAP-TEAD inhibitor that targets in-vitro vasculogenic mimicry in glioblastoma cells
Abstract
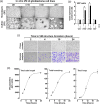
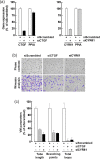
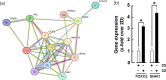
Glioblastoma (GBM) is a highly angiogenic malignancy of the central nervous system that resists standard antiangiogenic therapy, in part because of an alternative process to angiogenesis termed vasculogenic mimicry. Intricately linked to GBM, dysregulation of the Hippo signaling pathway leads to overexpression of YAP/TEAD and several downstream effectors involved in therapy resistance. Little is known about whether vasculogenic mimicry and the Hippo pathway intersect in the GBM chemoresistance phenotype. This study seeks to investigate the expression patterns of Hippo pathway regulators within clinically annotated GBM samples, examining their involvement in vitro regarding vasculogenic mimicry. In addition, it aims to assess the potential for pharmacological targeting of this pathway. In-silico analysis of the Hippo signaling members YAP1 , TEAD1 , AXL , NF2 , CTGF , and CYR61 transcript levels in low-grade GBM and GBM tumor tissues was done by Gene Expression Profiling Interactive Analysis. Gene expression was analyzed by real-time quantitative PCR from human U87, U118, U138, and U251 brain cancer cell lines and in clinically annotated brain tumor cDNA arrays. Transient gene silencing was performed with specific small interfering RNA. Vasculogenic mimicry was assessed using a Cultrex matrix, and three-dimensional capillary-like structures were analyzed with Wimasis. CYR61 and CTGF transcript levels were elevated in GBM tissues and were further induced when in-vitro vasculogenic mimicry was assessed. Silencing of CYR61 and CTGF , or treatment with a small-molecule TEAD inhibitor LM98 derived from flufenamic acid, inhibited vasculogenic mimicry. Silencing of SNAI1 and FOXC2 also altered vasculogenic mimicry and reduced CYR61 / CTGF levels. Pharmacological targeting of the Hippo pathway inhibits in-vitro vasculogenic mimicry. Unraveling the connections between the Hippo pathway and vasculogenic mimicry may pave the way for innovative therapeutic strategies.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
There are no conflicts of interest.
Figures




References
-
- GARD (Genetic and Rare Diseases Information Center). https://rarediseases.info.nih.gov/diseases/2491/glioblastoma. [Accessed 14 May 2024]
-
- Lassen U, Mau-Sørensen M, Skovgaard Poulsen H. Orphan drugs in glioblastoma multiforme: a review. Orphan Drugs Res Rev 2014; 4:83–91.
-
- Mariz S, Reese JH, Westermark K, Greene L, Goto T, Hoshino T, et al. . Worldwide collaboration for orphan drug designation. Nat Rev Drug Discov 2016; 15:440–441. - PubMed
-
- Di Nunno V, Franceschi E, Tosoni A, Di Battista M, Gatto L, Lamperini C, et al. . Treatment of recurrent glioblastoma: state-of-the-art and future perspectives. Expert Rev Anticancer Ther 2020; 20:785–795. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

