Guiding antibiotics towards their target using bacteriophage proteins
- PMID: 38902231
- PMCID: PMC11190222
- DOI: 10.1038/s41467-024-49603-4
Guiding antibiotics towards their target using bacteriophage proteins
Abstract
Novel therapeutic strategies against difficult-to-treat bacterial infections are desperately needed, and the faster and cheaper way to get them might be by repurposing existing antibiotics. Nanodelivery systems enhance the efficacy of antibiotics by guiding them to their targets, increasing the local concentration at the site of infection. While recently described nanodelivery systems are promising, they are generally not easy to adapt to different targets, and lack biocompatibility or specificity. Here, nanodelivery systems are created that source their targeting proteins from bacteriophages. Bacteriophage receptor-binding proteins and cell-wall binding domains are conjugated to nanoparticles, for the targeted delivery of rifampicin, imipenem, and ampicillin against bacterial pathogens. They show excellent specificity against their targets, and accumulate at the site of infection to deliver their antibiotic payload. Moreover, the nanodelivery systems suppress pathogen infections more effectively than 16 to 32-fold higher doses of free antibiotics. This study demonstrates that bacteriophage sourced targeting proteins are promising candidates to guide nanodelivery systems. Their specificity, availability, and biocompatibility make them great options to guide the antibiotic nanodelivery systems that are desperately needed to combat difficult-to-treat infections.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

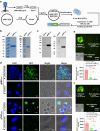

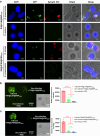
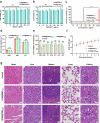
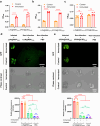


References
MeSH terms
Substances
Grants and funding
- 2022YFH0057/Sichuan Provincial Department of Science and Technology | Sichuan Province Science and Technology Support Program
- 2022YFH0062/Sichuan Provincial Department of Science and Technology | Sichuan Province Science and Technology Support Program
- 2022ZYD0068/Sichuan Provincial Department of Science and Technology | Sichuan Province Science and Technology Support Program
- 32102689/National Science Foundation of China | National Natural Science Foundation of China-Yunnan Joint Fund (NSFC-Yunnan Joint Fund)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

